| Full name: C-C motif chemokine ligand 11 | Alias Symbol: eotaxin|MGC22554 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 6356 | HGNC ID: HGNC:10610 | Ensembl Gene: ENSG00000172156 | OMIM ID: 601156 |
| Drug and gene relationship at DGIdb | |||
CCL11 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL11:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL11 | 6356 | 210133_at | -0.5687 | 0.6150 | |
| GSE20347 | CCL11 | 6356 | 210133_at | 0.1960 | 0.3639 | |
| GSE23400 | CCL11 | 6356 | 210133_at | 0.1208 | 0.1629 | |
| GSE26886 | CCL11 | 6356 | 210133_at | 0.4632 | 0.2449 | |
| GSE29001 | CCL11 | 6356 | 210133_at | -0.2742 | 0.1340 | |
| GSE38129 | CCL11 | 6356 | 210133_at | 0.0378 | 0.9263 | |
| GSE45670 | CCL11 | 6356 | 210133_at | -1.3684 | 0.0260 | |
| GSE53622 | CCL11 | 6356 | 90422 | -0.0279 | 0.9149 | |
| GSE53624 | CCL11 | 6356 | 90422 | -0.4196 | 0.0286 | |
| GSE63941 | CCL11 | 6356 | 210133_at | -5.9978 | 0.0000 | |
| GSE77861 | CCL11 | 6356 | 210133_at | 0.3368 | 0.3088 | |
| GSE97050 | CCL11 | 6356 | A_23_P66635 | 0.6478 | 0.2292 | |
| SRP007169 | CCL11 | 6356 | RNAseq | 1.1564 | 0.0410 | |
| SRP064894 | CCL11 | 6356 | RNAseq | -0.6248 | 0.1318 | |
| SRP133303 | CCL11 | 6356 | RNAseq | -0.3417 | 0.3163 | |
| SRP159526 | CCL11 | 6356 | RNAseq | 0.7482 | 0.1820 | |
| SRP193095 | CCL11 | 6356 | RNAseq | 1.0959 | 0.0094 | |
| SRP219564 | CCL11 | 6356 | RNAseq | 0.9211 | 0.2278 | |
| TCGA | CCL11 | 6356 | RNAseq | -0.3439 | 0.1362 |
Upregulated datasets: 2; Downregulated datasets: 2.
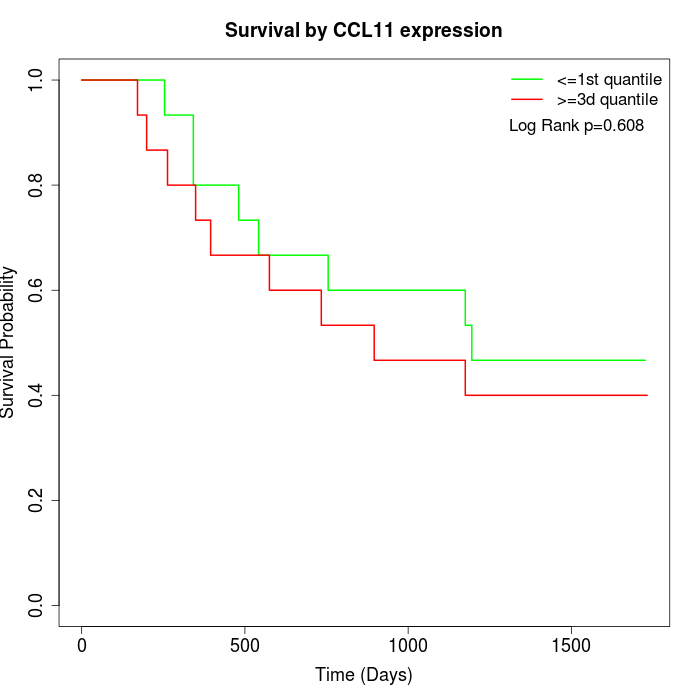
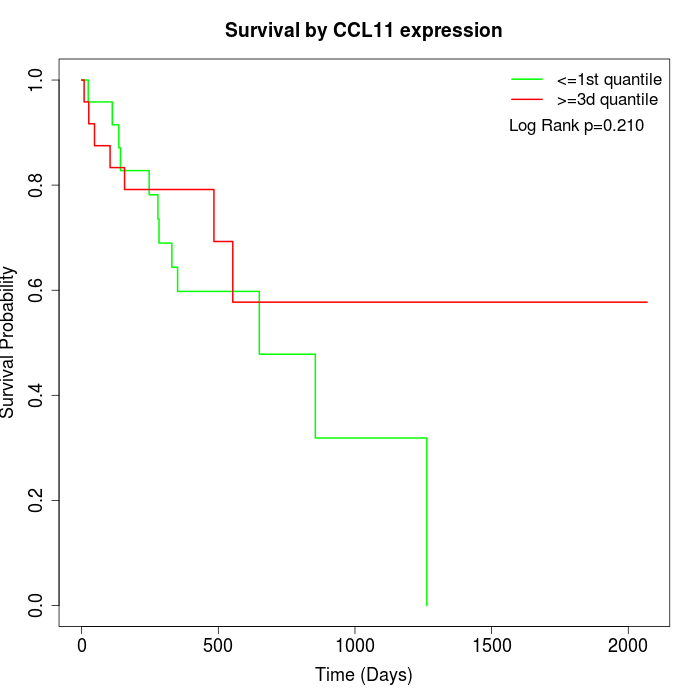
Survival by CCL11 expression:
Note: Click image to view full size file.
Copy number change of CCL11:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL11 | 6356 | 5 | 1 | 24 | |
| GSE20123 | CCL11 | 6356 | 5 | 1 | 24 | |
| GSE43470 | CCL11 | 6356 | 1 | 2 | 40 | |
| GSE46452 | CCL11 | 6356 | 34 | 0 | 25 | |
| GSE47630 | CCL11 | 6356 | 7 | 1 | 32 | |
| GSE54993 | CCL11 | 6356 | 3 | 3 | 64 | |
| GSE54994 | CCL11 | 6356 | 8 | 6 | 39 | |
| GSE60625 | CCL11 | 6356 | 4 | 0 | 7 | |
| GSE74703 | CCL11 | 6356 | 1 | 1 | 34 | |
| GSE74704 | CCL11 | 6356 | 3 | 1 | 16 | |
| TCGA | CCL11 | 6356 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of CCL11:
Generating mutation plots.
Highly correlated genes for CCL11:
Showing top 20/507 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL11 | ZNF469 | 0.765101 | 3 | 0 | 3 |
| CCL11 | DMBT1 | 0.745639 | 3 | 0 | 3 |
| CCL11 | CYTL1 | 0.729245 | 4 | 0 | 4 |
| CCL11 | MMP3 | 0.728129 | 3 | 0 | 3 |
| CCL11 | HAUS1 | 0.720246 | 3 | 0 | 3 |
| CCL11 | DPY19L1 | 0.717255 | 3 | 0 | 3 |
| CCL11 | PDE4B | 0.70682 | 6 | 0 | 6 |
| CCL11 | DEGS1 | 0.70491 | 3 | 0 | 3 |
| CCL11 | ANXA5 | 0.697394 | 3 | 0 | 3 |
| CCL11 | SPON1 | 0.696845 | 9 | 0 | 8 |
| CCL11 | CHST11 | 0.695571 | 3 | 0 | 3 |
| CCL11 | TCL1A | 0.694582 | 3 | 0 | 3 |
| CCL11 | NBPF20 | 0.693322 | 3 | 0 | 3 |
| CCL11 | LIMS1 | 0.69165 | 3 | 0 | 3 |
| CCL11 | CCDC102B | 0.687816 | 5 | 0 | 4 |
| CCL11 | PLEK | 0.684057 | 4 | 0 | 4 |
| CCL11 | COL15A1 | 0.678475 | 9 | 0 | 8 |
| CCL11 | ADAMTS12 | 0.676412 | 4 | 0 | 4 |
| CCL11 | ACKR1 | 0.675831 | 6 | 0 | 6 |
| CCL11 | CNRIP1 | 0.672039 | 9 | 0 | 7 |
For details and further investigation, click here