| Full name: ficolin 2 | Alias Symbol: P35|FCNL|EBP-37|ficolin-2 | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 2220 | HGNC ID: HGNC:3624 | Ensembl Gene: ENSG00000160339 | OMIM ID: 601624 |
| Drug and gene relationship at DGIdb | |||
Expression of FCN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FCN2 | 2220 | 208439_s_at | 0.0296 | 0.9438 | |
| GSE20347 | FCN2 | 2220 | 208439_s_at | 0.0621 | 0.3649 | |
| GSE23400 | FCN2 | 2220 | 208439_s_at | -0.1065 | 0.0057 | |
| GSE26886 | FCN2 | 2220 | 208439_s_at | 0.1665 | 0.1133 | |
| GSE29001 | FCN2 | 2220 | 208439_s_at | -0.1047 | 0.6985 | |
| GSE38129 | FCN2 | 2220 | 208439_s_at | -0.0847 | 0.4163 | |
| GSE45670 | FCN2 | 2220 | 208439_s_at | 0.0050 | 0.9689 | |
| GSE53622 | FCN2 | 2220 | 38227 | 0.0021 | 0.9816 | |
| GSE53624 | FCN2 | 2220 | 38227 | 0.1904 | 0.0276 | |
| GSE63941 | FCN2 | 2220 | 208439_s_at | 0.2085 | 0.0914 | |
| GSE77861 | FCN2 | 2220 | 208439_s_at | 0.0217 | 0.9009 | |
| GSE97050 | FCN2 | 2220 | A_23_P216756 | -0.0583 | 0.8267 |
Upregulated datasets: 0; Downregulated datasets: 0.
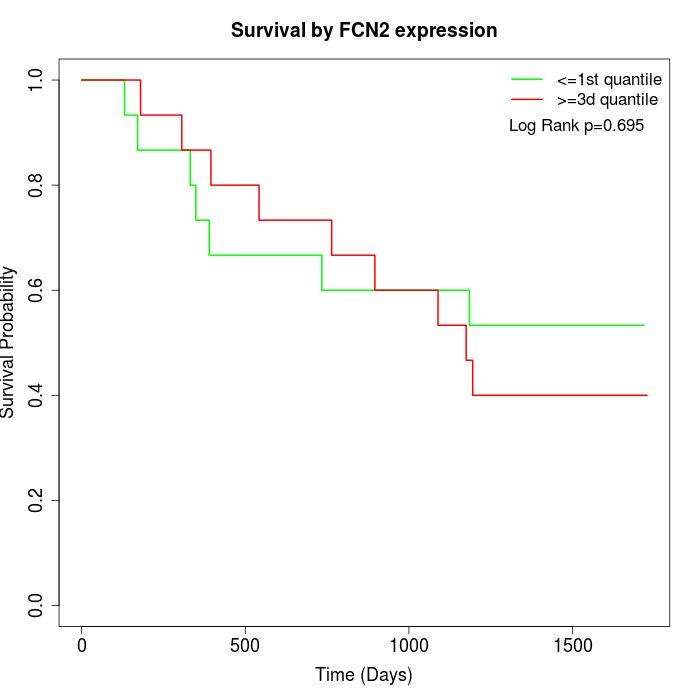
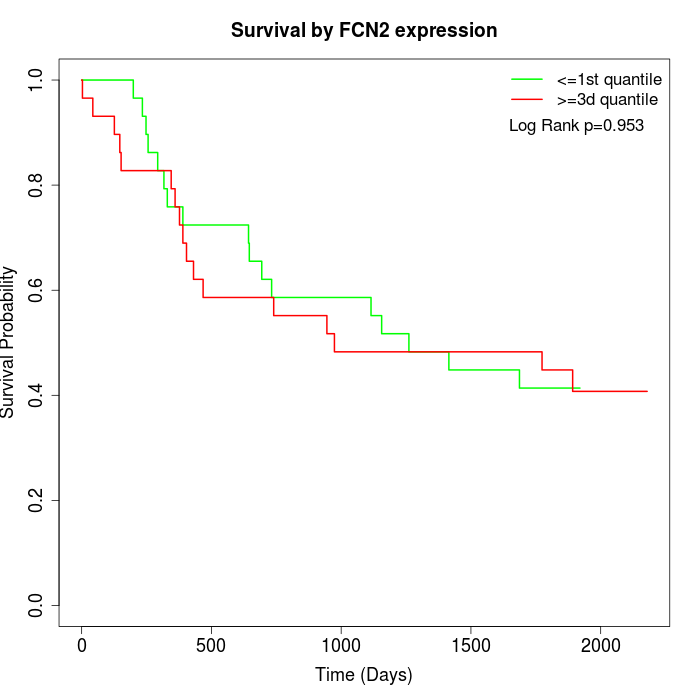
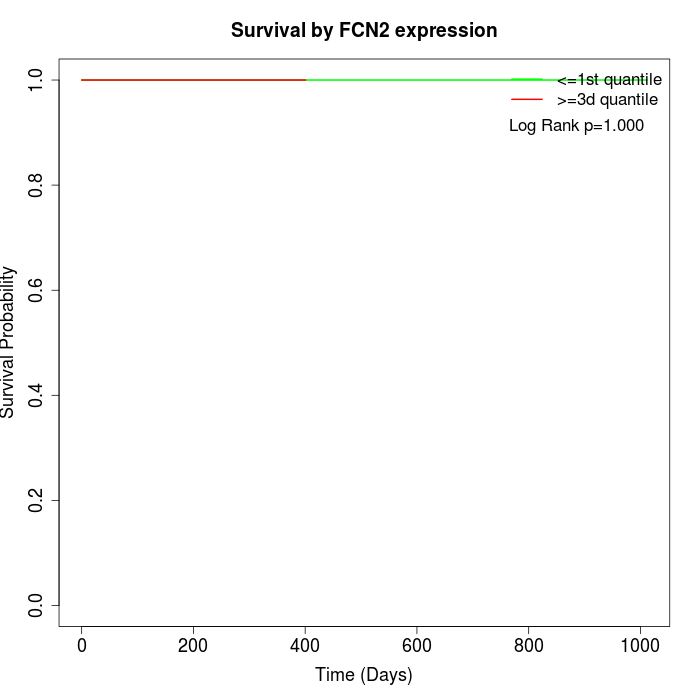
Survival by FCN2 expression:
Note: Click image to view full size file.
Copy number change of FCN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FCN2 | 2220 | 4 | 7 | 19 | |
| GSE20123 | FCN2 | 2220 | 4 | 7 | 19 | |
| GSE43470 | FCN2 | 2220 | 3 | 8 | 32 | |
| GSE46452 | FCN2 | 2220 | 6 | 13 | 40 | |
| GSE47630 | FCN2 | 2220 | 6 | 15 | 19 | |
| GSE54993 | FCN2 | 2220 | 3 | 3 | 64 | |
| GSE54994 | FCN2 | 2220 | 12 | 9 | 32 | |
| GSE60625 | FCN2 | 2220 | 0 | 0 | 11 | |
| GSE74703 | FCN2 | 2220 | 3 | 6 | 27 | |
| GSE74704 | FCN2 | 2220 | 2 | 5 | 13 | |
| TCGA | FCN2 | 2220 | 28 | 25 | 43 |
Total number of gains: 71; Total number of losses: 98; Total Number of normals: 319.
Somatic mutations of FCN2:
Generating mutation plots.
Highly correlated genes for FCN2:
Showing top 20/1133 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FCN2 | C3orf36 | 0.75714 | 4 | 0 | 4 |
| FCN2 | C1orf61 | 0.742156 | 4 | 0 | 4 |
| FCN2 | KIR3DL3 | 0.739146 | 5 | 0 | 4 |
| FCN2 | TBX4 | 0.738412 | 5 | 0 | 4 |
| FCN2 | PHF1 | 0.735119 | 3 | 0 | 3 |
| FCN2 | B3GALT1 | 0.735107 | 6 | 0 | 6 |
| FCN2 | RGSL1 | 0.728621 | 4 | 0 | 4 |
| FCN2 | CHRNA10 | 0.725804 | 4 | 0 | 4 |
| FCN2 | NTNG1 | 0.721291 | 3 | 0 | 3 |
| FCN2 | ADCYAP1R1 | 0.714171 | 3 | 0 | 3 |
| FCN2 | ATP8B3 | 0.710256 | 4 | 0 | 4 |
| FCN2 | CLSTN3 | 0.7076 | 5 | 0 | 4 |
| FCN2 | TMEM235 | 0.707059 | 3 | 0 | 3 |
| FCN2 | IFNA4 | 0.706705 | 6 | 0 | 5 |
| FCN2 | IL1RAPL2 | 0.705215 | 5 | 0 | 4 |
| FCN2 | SPAM1 | 0.701919 | 4 | 0 | 4 |
| FCN2 | RNF17 | 0.701498 | 4 | 0 | 4 |
| FCN2 | IFNA7 | 0.701228 | 4 | 0 | 4 |
| FCN2 | FFAR2 | 0.700892 | 3 | 0 | 3 |
| FCN2 | RNF123 | 0.699278 | 5 | 0 | 4 |
For details and further investigation, click here