| Full name: sperm adhesion molecule 1 | Alias Symbol: HYAL5|PH-20|SPAG15 | ||
| Type: protein-coding gene | Cytoband: 7q31 | ||
| Entrez ID: 6677 | HGNC ID: HGNC:11217 | Ensembl Gene: ENSG00000106304 | OMIM ID: 600930 |
| Drug and gene relationship at DGIdb | |||
Expression of SPAM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPAM1 | 6677 | 210536_s_at | -0.1476 | 0.5492 | |
| GSE20347 | SPAM1 | 6677 | 210536_s_at | -0.0076 | 0.9182 | |
| GSE23400 | SPAM1 | 6677 | 210536_s_at | -0.0402 | 0.1985 | |
| GSE26886 | SPAM1 | 6677 | 210536_s_at | -0.1130 | 0.3600 | |
| GSE29001 | SPAM1 | 6677 | 210536_s_at | -0.0861 | 0.5354 | |
| GSE38129 | SPAM1 | 6677 | 210536_s_at | -0.1053 | 0.1295 | |
| GSE45670 | SPAM1 | 6677 | 216989_at | 0.0479 | 0.6225 | |
| GSE53622 | SPAM1 | 6677 | 65808 | 0.1858 | 0.6061 | |
| GSE53624 | SPAM1 | 6677 | 65808 | 0.6871 | 0.0000 | |
| GSE63941 | SPAM1 | 6677 | 210536_s_at | 0.0080 | 0.9672 | |
| GSE77861 | SPAM1 | 6677 | 210536_s_at | -0.1203 | 0.2710 |
Upregulated datasets: 0; Downregulated datasets: 0.
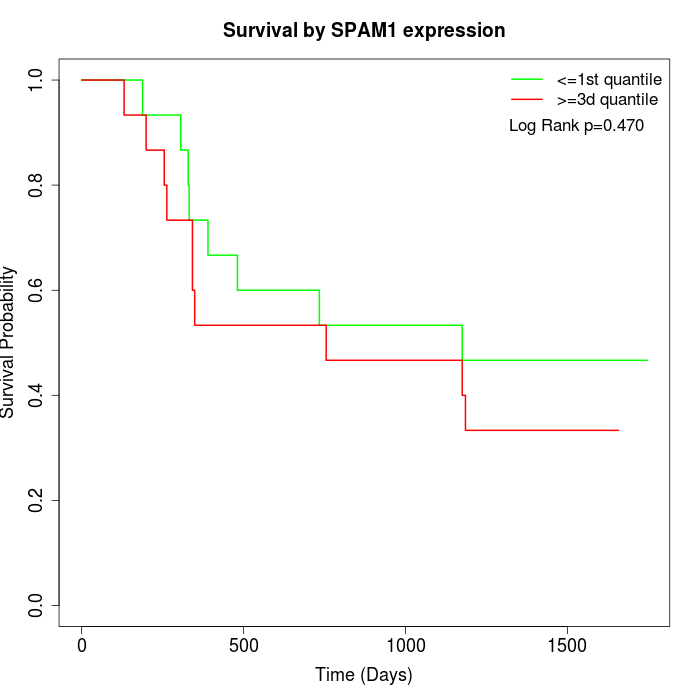
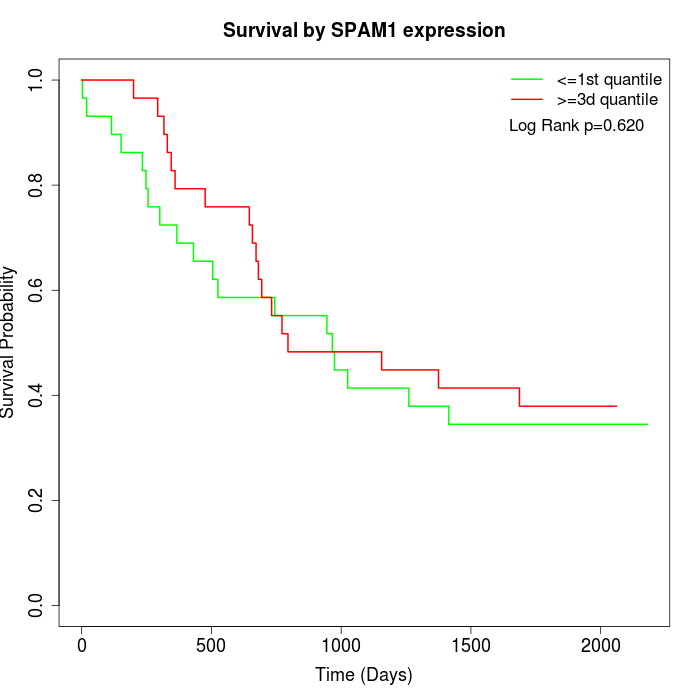
Survival by SPAM1 expression:
Note: Click image to view full size file.
Copy number change of SPAM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPAM1 | 6677 | 6 | 2 | 22 | |
| GSE20123 | SPAM1 | 6677 | 7 | 2 | 21 | |
| GSE43470 | SPAM1 | 6677 | 2 | 4 | 37 | |
| GSE46452 | SPAM1 | 6677 | 8 | 1 | 50 | |
| GSE47630 | SPAM1 | 6677 | 7 | 4 | 29 | |
| GSE54993 | SPAM1 | 6677 | 3 | 5 | 62 | |
| GSE54994 | SPAM1 | 6677 | 9 | 7 | 37 | |
| GSE60625 | SPAM1 | 6677 | 0 | 0 | 11 | |
| GSE74703 | SPAM1 | 6677 | 2 | 4 | 30 | |
| GSE74704 | SPAM1 | 6677 | 5 | 2 | 13 | |
| TCGA | SPAM1 | 6677 | 27 | 21 | 48 |
Total number of gains: 76; Total number of losses: 52; Total Number of normals: 360.
Somatic mutations of SPAM1:
Generating mutation plots.
Highly correlated genes for SPAM1:
Showing top 20/838 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPAM1 | DEFB108B | 0.737555 | 3 | 0 | 3 |
| SPAM1 | PGC | 0.71168 | 3 | 0 | 3 |
| SPAM1 | FCN2 | 0.701919 | 4 | 0 | 4 |
| SPAM1 | NXPE4 | 0.682678 | 4 | 0 | 4 |
| SPAM1 | CCL16 | 0.678351 | 4 | 0 | 4 |
| SPAM1 | LGALS14 | 0.672523 | 5 | 0 | 5 |
| SPAM1 | CGA | 0.672205 | 4 | 0 | 3 |
| SPAM1 | SLC10A2 | 0.670734 | 4 | 0 | 4 |
| SPAM1 | TCP10L | 0.667948 | 5 | 0 | 5 |
| SPAM1 | KRT38 | 0.667683 | 4 | 0 | 3 |
| SPAM1 | FEV | 0.667029 | 7 | 0 | 6 |
| SPAM1 | LINC00857 | 0.665692 | 3 | 0 | 3 |
| SPAM1 | LINC00868 | 0.665105 | 3 | 0 | 3 |
| SPAM1 | CLEC4M | 0.664281 | 4 | 0 | 4 |
| SPAM1 | FSCN3 | 0.661222 | 6 | 0 | 5 |
| SPAM1 | PSG9 | 0.659807 | 5 | 0 | 4 |
| SPAM1 | EPOR | 0.659225 | 6 | 0 | 6 |
| SPAM1 | EDA2R | 0.659081 | 6 | 0 | 5 |
| SPAM1 | BTNL3 | 0.658944 | 5 | 0 | 4 |
| SPAM1 | KIR2DL5A | 0.657974 | 4 | 0 | 4 |
For details and further investigation, click here