| Full name: regulator of G protein signaling like 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q25.3 | ||
| Entrez ID: 353299 | HGNC ID: HGNC:18636 | Ensembl Gene: ENSG00000121446 | OMIM ID: 611012 |
| Drug and gene relationship at DGIdb | |||
Expression of RGSL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGSL1 | 353299 | 211446_at | -0.0455 | 0.9024 | |
| GSE20347 | RGSL1 | 353299 | 211446_at | -0.1186 | 0.2403 | |
| GSE23400 | RGSL1 | 353299 | 211446_at | -0.0969 | 0.0050 | |
| GSE26886 | RGSL1 | 353299 | 211446_at | -0.1585 | 0.1977 | |
| GSE29001 | RGSL1 | 353299 | 211446_at | -0.0975 | 0.5726 | |
| GSE38129 | RGSL1 | 353299 | 211446_at | -0.1392 | 0.2933 | |
| GSE45670 | RGSL1 | 353299 | 211446_at | 0.0097 | 0.9385 | |
| GSE53622 | RGSL1 | 353299 | 66998 | 0.4237 | 0.0050 | |
| GSE53624 | RGSL1 | 353299 | 66998 | 0.9030 | 0.0000 | |
| GSE63941 | RGSL1 | 353299 | 211446_at | -0.0111 | 0.9414 | |
| GSE77861 | RGSL1 | 353299 | 211446_at | -0.2281 | 0.0104 | |
| TCGA | RGSL1 | 353299 | RNAseq | 3.6905 | 0.0006 |
Upregulated datasets: 1; Downregulated datasets: 0.
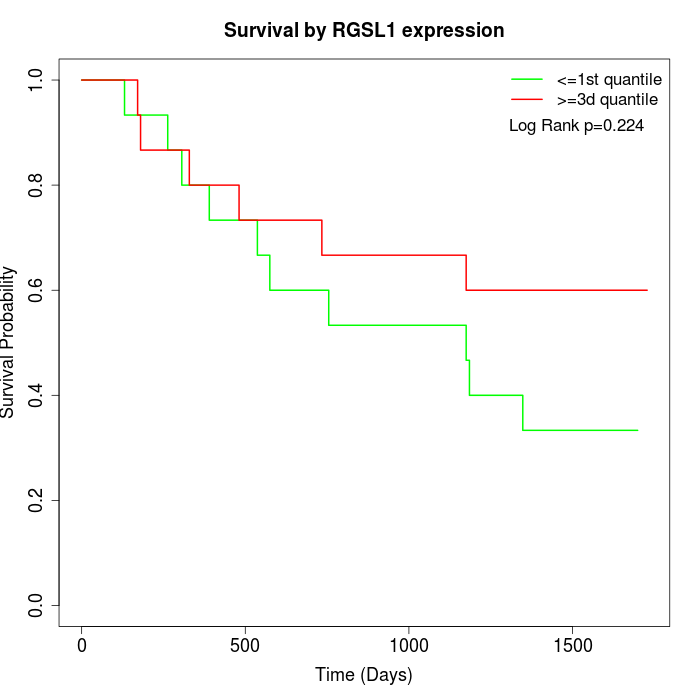
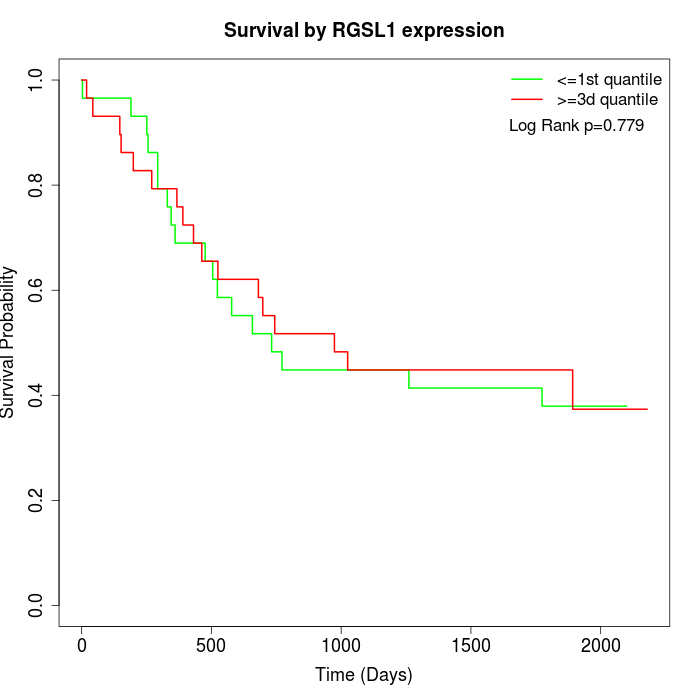
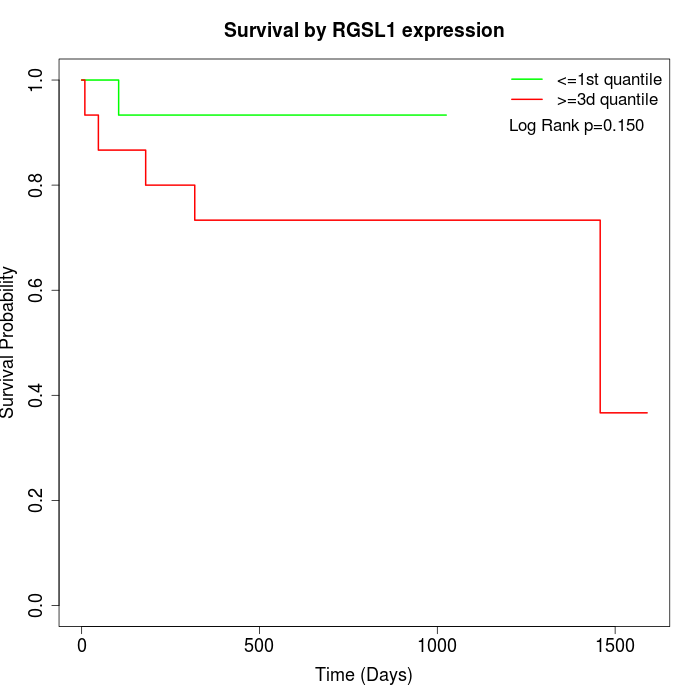
Survival by RGSL1 expression:
Note: Click image to view full size file.
Copy number change of RGSL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGSL1 | 353299 | 12 | 0 | 18 | |
| GSE20123 | RGSL1 | 353299 | 12 | 0 | 18 | |
| GSE43470 | RGSL1 | 353299 | 7 | 1 | 35 | |
| GSE46452 | RGSL1 | 353299 | 3 | 1 | 55 | |
| GSE47630 | RGSL1 | 353299 | 14 | 0 | 26 | |
| GSE54993 | RGSL1 | 353299 | 0 | 6 | 64 | |
| GSE54994 | RGSL1 | 353299 | 15 | 0 | 38 | |
| GSE60625 | RGSL1 | 353299 | 0 | 0 | 11 | |
| GSE74703 | RGSL1 | 353299 | 7 | 1 | 28 | |
| GSE74704 | RGSL1 | 353299 | 5 | 0 | 15 | |
| TCGA | RGSL1 | 353299 | 40 | 3 | 53 |
Total number of gains: 115; Total number of losses: 12; Total Number of normals: 361.
Somatic mutations of RGSL1:
Generating mutation plots.
Highly correlated genes for RGSL1:
Showing top 20/888 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGSL1 | FCN2 | 0.728621 | 4 | 0 | 4 |
| RGSL1 | MYOD1 | 0.726669 | 3 | 0 | 3 |
| RGSL1 | PHLDB1 | 0.722541 | 5 | 0 | 5 |
| RGSL1 | ITGB3 | 0.72207 | 3 | 0 | 3 |
| RGSL1 | CYP2D6 | 0.720491 | 4 | 0 | 4 |
| RGSL1 | NPAP1 | 0.718187 | 4 | 0 | 4 |
| RGSL1 | EDA2R | 0.715999 | 5 | 0 | 4 |
| RGSL1 | MTAP | 0.710659 | 4 | 0 | 4 |
| RGSL1 | MUC8 | 0.699852 | 5 | 0 | 5 |
| RGSL1 | OR3A3 | 0.698803 | 4 | 0 | 4 |
| RGSL1 | NGB | 0.695564 | 4 | 0 | 4 |
| RGSL1 | KCNJ10 | 0.694331 | 5 | 0 | 5 |
| RGSL1 | CTRB2 | 0.690227 | 3 | 0 | 3 |
| RGSL1 | GABRB2 | 0.686747 | 4 | 0 | 4 |
| RGSL1 | CER1 | 0.681254 | 6 | 0 | 5 |
| RGSL1 | PIWIL2 | 0.680337 | 5 | 0 | 5 |
| RGSL1 | CPNE6 | 0.679878 | 5 | 0 | 5 |
| RGSL1 | CCR9 | 0.679792 | 5 | 0 | 5 |
| RGSL1 | DGCR5 | 0.678608 | 4 | 0 | 4 |
| RGSL1 | ADRB3 | 0.678304 | 5 | 0 | 4 |
For details and further investigation, click here