| Full name: FES proto-oncogene, tyrosine kinase | Alias Symbol: FPS | ||
| Type: protein-coding gene | Cytoband: 15q26.1 | ||
| Entrez ID: 2242 | HGNC ID: HGNC:3657 | Ensembl Gene: ENSG00000182511 | OMIM ID: 190030 |
| Drug and gene relationship at DGIdb | |||
FES involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of FES:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FES | 2242 | 205418_at | -0.0042 | 0.9942 | |
| GSE20347 | FES | 2242 | 205418_at | 0.1520 | 0.1659 | |
| GSE23400 | FES | 2242 | 205418_at | -0.1203 | 0.0032 | |
| GSE26886 | FES | 2242 | 205418_at | 0.0795 | 0.5984 | |
| GSE29001 | FES | 2242 | 205418_at | 0.0657 | 0.6184 | |
| GSE38129 | FES | 2242 | 205418_at | -0.0842 | 0.6218 | |
| GSE45670 | FES | 2242 | 205418_at | -0.0314 | 0.8361 | |
| GSE53622 | FES | 2242 | 47652 | -0.2025 | 0.0032 | |
| GSE53624 | FES | 2242 | 47652 | -0.3282 | 0.0000 | |
| GSE63941 | FES | 2242 | 205418_at | -0.2857 | 0.7316 | |
| GSE77861 | FES | 2242 | 205418_at | -0.1059 | 0.3685 | |
| GSE97050 | FES | 2242 | A_23_P14769 | 0.2592 | 0.3142 | |
| SRP007169 | FES | 2242 | RNAseq | 1.1653 | 0.0543 | |
| SRP064894 | FES | 2242 | RNAseq | 0.6989 | 0.0333 | |
| SRP133303 | FES | 2242 | RNAseq | -0.2992 | 0.1179 | |
| SRP159526 | FES | 2242 | RNAseq | 0.1130 | 0.7597 | |
| SRP193095 | FES | 2242 | RNAseq | 0.5453 | 0.0046 | |
| SRP219564 | FES | 2242 | RNAseq | 0.4815 | 0.2594 | |
| TCGA | FES | 2242 | RNAseq | -0.1637 | 0.1991 |
Upregulated datasets: 0; Downregulated datasets: 0.
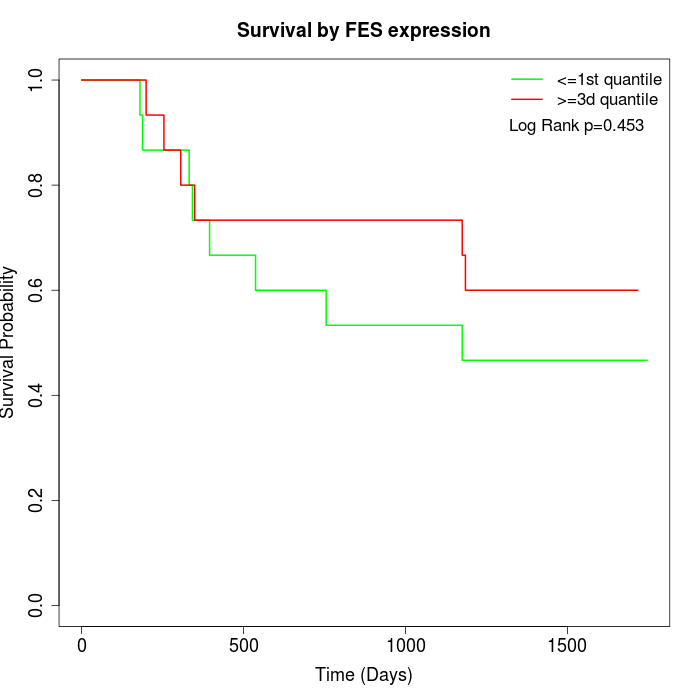
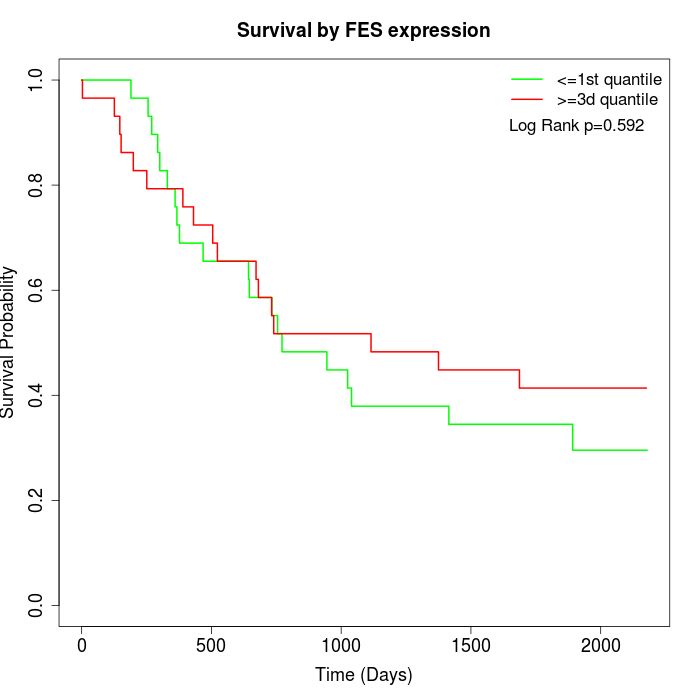
Survival by FES expression:
Note: Click image to view full size file.
Copy number change of FES:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FES | 2242 | 8 | 2 | 20 | |
| GSE20123 | FES | 2242 | 8 | 2 | 20 | |
| GSE43470 | FES | 2242 | 5 | 5 | 33 | |
| GSE46452 | FES | 2242 | 3 | 7 | 49 | |
| GSE47630 | FES | 2242 | 8 | 11 | 21 | |
| GSE54993 | FES | 2242 | 4 | 6 | 60 | |
| GSE54994 | FES | 2242 | 7 | 7 | 39 | |
| GSE60625 | FES | 2242 | 4 | 0 | 7 | |
| GSE74703 | FES | 2242 | 4 | 4 | 28 | |
| GSE74704 | FES | 2242 | 4 | 2 | 14 | |
| TCGA | FES | 2242 | 18 | 12 | 66 |
Total number of gains: 73; Total number of losses: 58; Total Number of normals: 357.
Somatic mutations of FES:
Generating mutation plots.
Highly correlated genes for FES:
Showing top 20/59 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FES | CASS4 | 0.799058 | 3 | 0 | 3 |
| FES | S1PR4 | 0.71162 | 3 | 0 | 3 |
| FES | MAGEE1 | 0.703544 | 3 | 0 | 3 |
| FES | BAHCC1 | 0.682384 | 3 | 0 | 3 |
| FES | PVALB | 0.679653 | 3 | 0 | 3 |
| FES | PTGIR | 0.674518 | 4 | 0 | 3 |
| FES | SNAI1 | 0.664427 | 4 | 0 | 3 |
| FES | SLC25A48 | 0.645027 | 3 | 0 | 3 |
| FES | TBXAS1 | 0.64463 | 4 | 0 | 3 |
| FES | PHOSPHO1 | 0.638874 | 4 | 0 | 4 |
| FES | CABP4 | 0.634831 | 3 | 0 | 3 |
| FES | STX1B | 0.626737 | 3 | 0 | 3 |
| FES | KCNE4 | 0.620435 | 3 | 0 | 3 |
| FES | OLIG3 | 0.612288 | 3 | 0 | 3 |
| FES | VASH2 | 0.608306 | 4 | 0 | 3 |
| FES | FCER2 | 0.596931 | 3 | 0 | 3 |
| FES | HLX | 0.595348 | 5 | 0 | 3 |
| FES | BTK | 0.592925 | 4 | 0 | 3 |
| FES | HSPB2 | 0.59231 | 3 | 0 | 3 |
| FES | CCL27 | 0.591216 | 4 | 0 | 3 |
For details and further investigation, click here