| Full name: C-C motif chemokine ligand 27 | Alias Symbol: ALP|ILC|CTACK|skinkine|ESkine|PESKY|CTAK | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 10850 | HGNC ID: HGNC:10626 | Ensembl Gene: ENSG00000213927 | OMIM ID: 604833 |
| Drug and gene relationship at DGIdb | |||
CCL27 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL27:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL27 | 10850 | 207955_at | -0.0068 | 0.9879 | |
| GSE20347 | CCL27 | 10850 | 207955_at | 0.1781 | 0.0088 | |
| GSE23400 | CCL27 | 10850 | 207955_at | -0.0504 | 0.0465 | |
| GSE26886 | CCL27 | 10850 | 207955_at | 0.2901 | 0.0712 | |
| GSE29001 | CCL27 | 10850 | 207955_at | -0.0047 | 0.9745 | |
| GSE38129 | CCL27 | 10850 | 207955_at | 0.0965 | 0.1560 | |
| GSE45670 | CCL27 | 10850 | 207955_at | 0.2060 | 0.2547 | |
| GSE63941 | CCL27 | 10850 | 207955_at | 0.1529 | 0.3810 | |
| GSE77861 | CCL27 | 10850 | 207955_at | -0.1676 | 0.3647 | |
| GSE97050 | CCL27 | 10850 | A_23_P135248 | 0.0231 | 0.9216 | |
| TCGA | CCL27 | 10850 | RNAseq | 2.2807 | 0.1275 |
Upregulated datasets: 0; Downregulated datasets: 0.
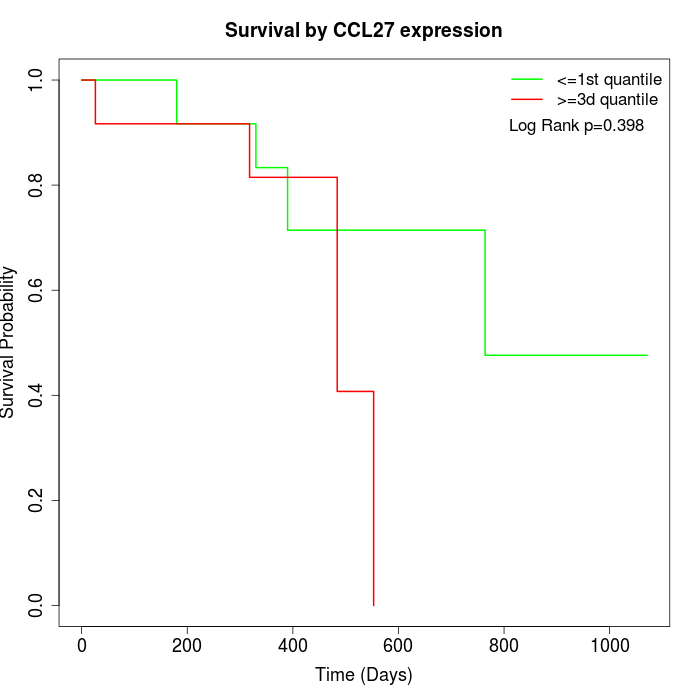
Survival by CCL27 expression:
Note: Click image to view full size file.
Copy number change of CCL27:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL27 | 10850 | 4 | 13 | 13 | |
| GSE20123 | CCL27 | 10850 | 4 | 13 | 13 | |
| GSE43470 | CCL27 | 10850 | 3 | 10 | 30 | |
| GSE46452 | CCL27 | 10850 | 6 | 15 | 38 | |
| GSE47630 | CCL27 | 10850 | 1 | 20 | 19 | |
| GSE54993 | CCL27 | 10850 | 6 | 0 | 64 | |
| GSE54994 | CCL27 | 10850 | 6 | 12 | 35 | |
| GSE60625 | CCL27 | 10850 | 0 | 0 | 11 | |
| GSE74703 | CCL27 | 10850 | 2 | 7 | 27 | |
| GSE74704 | CCL27 | 10850 | 0 | 11 | 9 | |
| TCGA | CCL27 | 10850 | 17 | 44 | 35 |
Total number of gains: 49; Total number of losses: 145; Total Number of normals: 294.
Somatic mutations of CCL27:
Generating mutation plots.
Highly correlated genes for CCL27:
Showing top 20/574 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL27 | FSCN2 | 0.754936 | 4 | 0 | 4 |
| CCL27 | CPN1 | 0.745011 | 4 | 0 | 4 |
| CCL27 | ALDH8A1 | 0.72884 | 3 | 0 | 3 |
| CCL27 | PRG3 | 0.728759 | 3 | 0 | 3 |
| CCL27 | SLC8A3 | 0.724951 | 3 | 0 | 3 |
| CCL27 | CHST10 | 0.718193 | 3 | 0 | 3 |
| CCL27 | ZFPM1 | 0.714702 | 3 | 0 | 3 |
| CCL27 | FFAR1 | 0.713093 | 3 | 0 | 3 |
| CCL27 | TSNAXIP1 | 0.712928 | 3 | 0 | 3 |
| CCL27 | CLSTN3 | 0.710243 | 3 | 0 | 3 |
| CCL27 | C7orf61 | 0.706483 | 3 | 0 | 3 |
| CCL27 | ESPNL | 0.703358 | 3 | 0 | 3 |
| CCL27 | FOLH1 | 0.697433 | 4 | 0 | 4 |
| CCL27 | GPSM1 | 0.696779 | 3 | 0 | 3 |
| CCL27 | MATN1 | 0.694696 | 3 | 0 | 3 |
| CCL27 | HHIP | 0.694683 | 4 | 0 | 3 |
| CCL27 | CALCR | 0.690592 | 4 | 0 | 4 |
| CCL27 | LRTM2 | 0.685342 | 4 | 0 | 3 |
| CCL27 | NXNL1 | 0.683857 | 4 | 0 | 4 |
| CCL27 | LMF1 | 0.6838 | 3 | 0 | 3 |
For details and further investigation, click here