| Full name: phosphoethanolamine/phosphocholine phosphatase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q21.32 | ||
| Entrez ID: 162466 | HGNC ID: HGNC:16815 | Ensembl Gene: ENSG00000173868 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of PHOSPHO1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PHOSPHO1 | 162466 | 236218_at | 0.0711 | 0.8213 | |
| GSE26886 | PHOSPHO1 | 162466 | 236218_at | 0.1029 | 0.3928 | |
| GSE45670 | PHOSPHO1 | 162466 | 236218_at | 0.0108 | 0.9148 | |
| GSE53622 | PHOSPHO1 | 162466 | 6364 | -0.1117 | 0.2866 | |
| GSE53624 | PHOSPHO1 | 162466 | 6364 | -0.4276 | 0.0001 | |
| GSE63941 | PHOSPHO1 | 162466 | 236218_at | 0.4298 | 0.0003 | |
| GSE77861 | PHOSPHO1 | 162466 | 236218_at | -0.2096 | 0.1401 | |
| GSE97050 | PHOSPHO1 | 162466 | A_33_P3332135 | -0.0509 | 0.8568 | |
| SRP133303 | PHOSPHO1 | 162466 | RNAseq | -0.4754 | 0.0164 | |
| SRP159526 | PHOSPHO1 | 162466 | RNAseq | 1.8629 | 0.0005 | |
| SRP193095 | PHOSPHO1 | 162466 | RNAseq | 0.1225 | 0.5731 | |
| SRP219564 | PHOSPHO1 | 162466 | RNAseq | -0.5898 | 0.3664 | |
| TCGA | PHOSPHO1 | 162466 | RNAseq | 0.2198 | 0.5034 |
Upregulated datasets: 1; Downregulated datasets: 0.
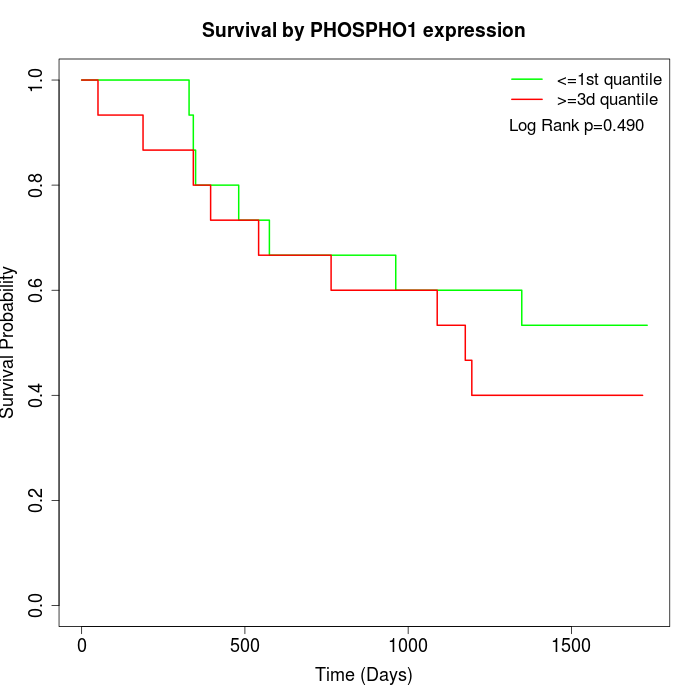
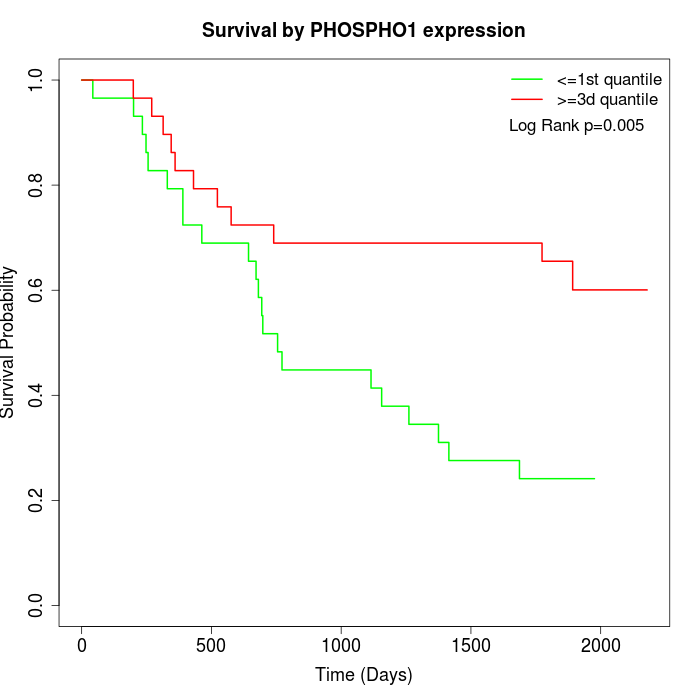
Survival by PHOSPHO1 expression:
Note: Click image to view full size file.
Copy number change of PHOSPHO1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PHOSPHO1 | 162466 | 6 | 2 | 22 | |
| GSE20123 | PHOSPHO1 | 162466 | 6 | 2 | 22 | |
| GSE43470 | PHOSPHO1 | 162466 | 3 | 2 | 38 | |
| GSE46452 | PHOSPHO1 | 162466 | 32 | 0 | 27 | |
| GSE47630 | PHOSPHO1 | 162466 | 9 | 0 | 31 | |
| GSE54993 | PHOSPHO1 | 162466 | 2 | 4 | 64 | |
| GSE54994 | PHOSPHO1 | 162466 | 10 | 6 | 37 | |
| GSE60625 | PHOSPHO1 | 162466 | 4 | 0 | 7 | |
| GSE74703 | PHOSPHO1 | 162466 | 3 | 2 | 31 | |
| GSE74704 | PHOSPHO1 | 162466 | 4 | 1 | 15 | |
| TCGA | PHOSPHO1 | 162466 | 27 | 7 | 62 |
Total number of gains: 106; Total number of losses: 26; Total Number of normals: 356.
Somatic mutations of PHOSPHO1:
Generating mutation plots.
Highly correlated genes for PHOSPHO1:
Showing top 20/330 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PHOSPHO1 | PRRG3 | 0.815679 | 3 | 0 | 3 |
| PHOSPHO1 | SLC6A18 | 0.813523 | 3 | 0 | 3 |
| PHOSPHO1 | ZC3H10 | 0.762648 | 3 | 0 | 3 |
| PHOSPHO1 | BARHL2 | 0.752544 | 3 | 0 | 3 |
| PHOSPHO1 | PNPLA7 | 0.743921 | 3 | 0 | 3 |
| PHOSPHO1 | C20orf173 | 0.743547 | 3 | 0 | 3 |
| PHOSPHO1 | CALCR | 0.742853 | 3 | 0 | 3 |
| PHOSPHO1 | MRGPRE | 0.742462 | 3 | 0 | 3 |
| PHOSPHO1 | PHGR1 | 0.741414 | 3 | 0 | 3 |
| PHOSPHO1 | EFCAB13 | 0.735481 | 3 | 0 | 3 |
| PHOSPHO1 | C4BPA | 0.733296 | 3 | 0 | 3 |
| PHOSPHO1 | WFIKKN2 | 0.724431 | 4 | 0 | 3 |
| PHOSPHO1 | ZMYND10 | 0.724184 | 4 | 0 | 4 |
| PHOSPHO1 | KCNJ13 | 0.72415 | 5 | 0 | 5 |
| PHOSPHO1 | RNF123 | 0.723733 | 3 | 0 | 3 |
| PHOSPHO1 | SPACA3 | 0.717304 | 4 | 0 | 4 |
| PHOSPHO1 | OR13G1 | 0.715794 | 3 | 0 | 3 |
| PHOSPHO1 | RUFY4 | 0.715145 | 3 | 0 | 3 |
| PHOSPHO1 | C11orf21 | 0.71468 | 3 | 0 | 3 |
| PHOSPHO1 | KANK3 | 0.711613 | 4 | 0 | 4 |
For details and further investigation, click here