| Full name: potassium calcium-activated channel subfamily M regulatory beta subunit 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q26.32 | ||
| Entrez ID: 10242 | HGNC ID: HGNC:6286 | Ensembl Gene: ENSG00000197584 | OMIM ID: 605214 |
| Drug and gene relationship at DGIdb | |||
KCNMB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04270 | Vascular smooth muscle contraction |
Expression of KCNMB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNMB2 | 10242 | 221097_s_at | -0.2592 | 0.2915 | |
| GSE20347 | KCNMB2 | 10242 | 221097_s_at | 0.0419 | 0.5313 | |
| GSE23400 | KCNMB2 | 10242 | 221097_s_at | -0.1337 | 0.0018 | |
| GSE26886 | KCNMB2 | 10242 | 221097_s_at | 0.0772 | 0.5384 | |
| GSE29001 | KCNMB2 | 10242 | 221097_s_at | -0.2053 | 0.3700 | |
| GSE38129 | KCNMB2 | 10242 | 221097_s_at | -0.1080 | 0.1950 | |
| GSE45670 | KCNMB2 | 10242 | 221097_s_at | -0.0303 | 0.7219 | |
| GSE53622 | KCNMB2 | 10242 | 68474 | 1.0575 | 0.0000 | |
| GSE53624 | KCNMB2 | 10242 | 68474 | 1.0632 | 0.0000 | |
| GSE63941 | KCNMB2 | 10242 | 221097_s_at | 0.0508 | 0.8298 | |
| GSE77861 | KCNMB2 | 10242 | 221097_s_at | -0.0779 | 0.5801 | |
| GSE97050 | KCNMB2 | 10242 | A_33_P3395191 | 0.5050 | 0.2451 | |
| SRP007169 | KCNMB2 | 10242 | RNAseq | 0.5138 | 0.4995 | |
| SRP133303 | KCNMB2 | 10242 | RNAseq | 1.9639 | 0.0000 | |
| SRP159526 | KCNMB2 | 10242 | RNAseq | 2.2512 | 0.0028 | |
| TCGA | KCNMB2 | 10242 | RNAseq | -1.5451 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 1.
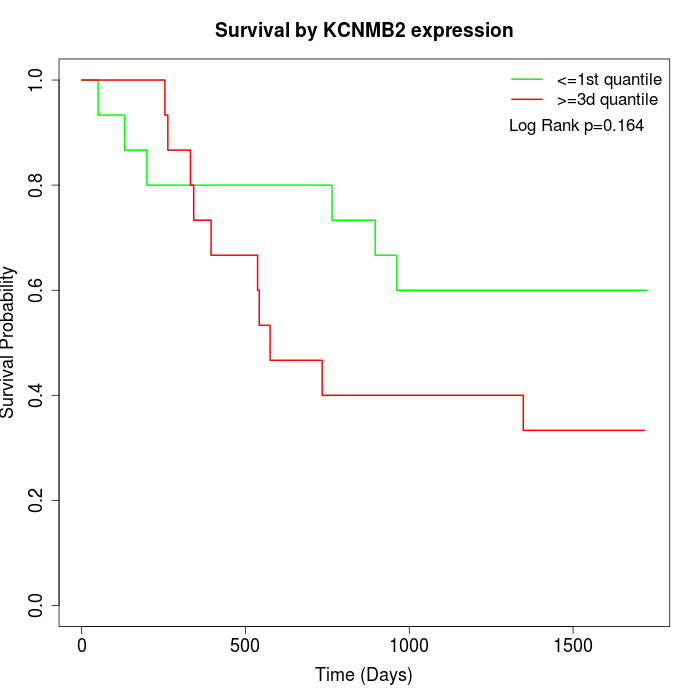
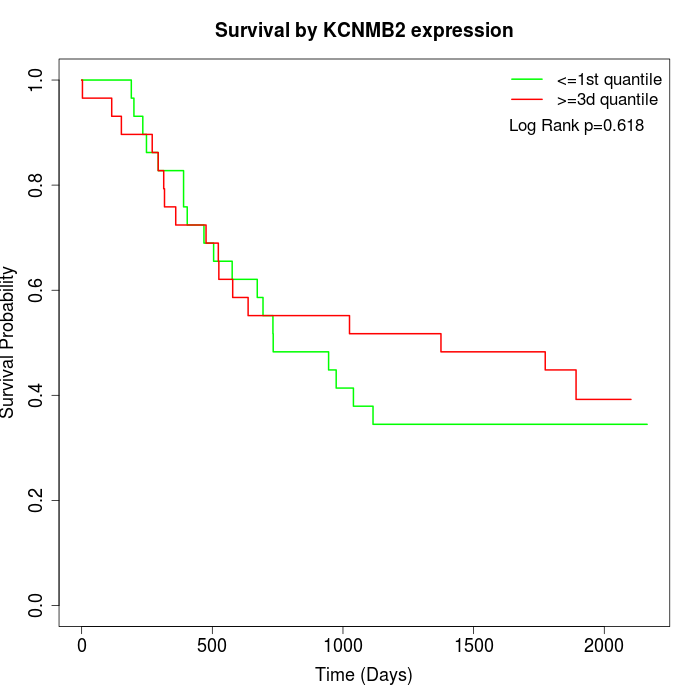
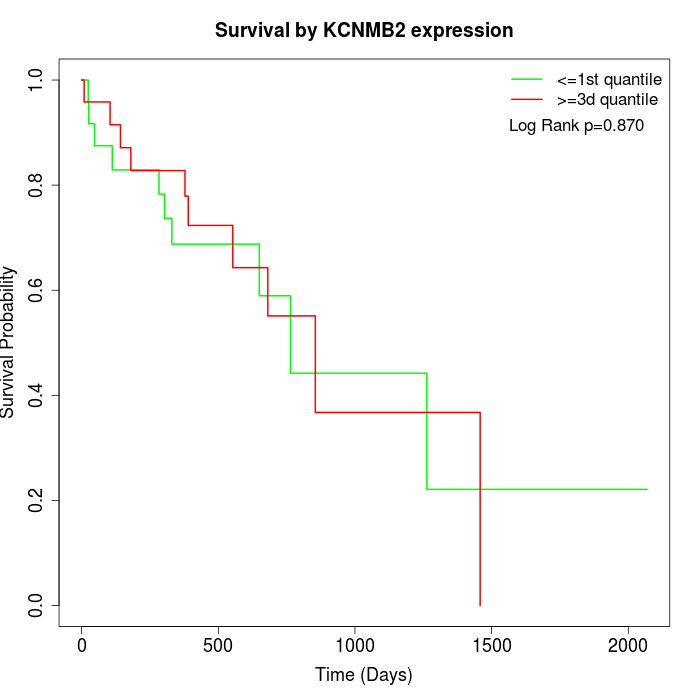
Survival by KCNMB2 expression:
Note: Click image to view full size file.
Copy number change of KCNMB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNMB2 | 10242 | 25 | 0 | 5 | |
| GSE20123 | KCNMB2 | 10242 | 25 | 0 | 5 | |
| GSE43470 | KCNMB2 | 10242 | 25 | 0 | 18 | |
| GSE46452 | KCNMB2 | 10242 | 19 | 1 | 39 | |
| GSE47630 | KCNMB2 | 10242 | 27 | 2 | 11 | |
| GSE54993 | KCNMB2 | 10242 | 1 | 18 | 51 | |
| GSE54994 | KCNMB2 | 10242 | 41 | 0 | 12 | |
| GSE60625 | KCNMB2 | 10242 | 0 | 6 | 5 | |
| GSE74703 | KCNMB2 | 10242 | 21 | 0 | 15 | |
| GSE74704 | KCNMB2 | 10242 | 16 | 0 | 4 | |
| TCGA | KCNMB2 | 10242 | 78 | 0 | 18 |
Total number of gains: 278; Total number of losses: 27; Total Number of normals: 183.
Somatic mutations of KCNMB2:
Generating mutation plots.
Highly correlated genes for KCNMB2:
Showing top 20/895 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNMB2 | KRTAP4-8 | 0.76967 | 3 | 0 | 3 |
| KCNMB2 | PGC | 0.751193 | 3 | 0 | 3 |
| KCNMB2 | PPM1N | 0.746439 | 3 | 0 | 3 |
| KCNMB2 | LACRT | 0.737381 | 3 | 0 | 3 |
| KCNMB2 | GSTM5 | 0.729128 | 4 | 0 | 4 |
| KCNMB2 | TLR8 | 0.726532 | 3 | 0 | 3 |
| KCNMB2 | MYO15A | 0.723875 | 4 | 0 | 4 |
| KCNMB2 | RASGRF2 | 0.721481 | 3 | 0 | 3 |
| KCNMB2 | CCKAR | 0.716287 | 4 | 0 | 4 |
| KCNMB2 | GPLD1 | 0.712469 | 4 | 0 | 4 |
| KCNMB2 | SLC10A2 | 0.712171 | 4 | 0 | 3 |
| KCNMB2 | CGA | 0.710388 | 5 | 0 | 5 |
| KCNMB2 | HCRT | 0.709441 | 5 | 0 | 5 |
| KCNMB2 | ABCC6 | 0.705292 | 3 | 0 | 3 |
| KCNMB2 | AICDA | 0.704766 | 4 | 0 | 3 |
| KCNMB2 | GABRA1 | 0.701094 | 5 | 0 | 4 |
| KCNMB2 | DND1 | 0.697824 | 3 | 0 | 3 |
| KCNMB2 | FMNL1 | 0.697791 | 3 | 0 | 3 |
| KCNMB2 | CCDC87 | 0.69328 | 3 | 0 | 3 |
| KCNMB2 | TESC | 0.693002 | 4 | 0 | 3 |
For details and further investigation, click here