| Full name: Rh associated glycoprotein | Alias Symbol: RH50A|CD241|SLC42A1 | ||
| Type: protein-coding gene | Cytoband: 6p12.3 | ||
| Entrez ID: 6005 | HGNC ID: HGNC:10006 | Ensembl Gene: ENSG00000112077 | OMIM ID: 180297 |
| Drug and gene relationship at DGIdb | |||
Expression of RHAG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RHAG | 6005 | 206146_s_at | 0.0966 | 0.6384 | |
| GSE20347 | RHAG | 6005 | 206146_s_at | 0.0632 | 0.4491 | |
| GSE23400 | RHAG | 6005 | 211254_x_at | -0.2842 | 0.0000 | |
| GSE26886 | RHAG | 6005 | 206146_s_at | 0.3331 | 0.0057 | |
| GSE29001 | RHAG | 6005 | 211254_x_at | -0.1557 | 0.4174 | |
| GSE38129 | RHAG | 6005 | 206146_s_at | -0.0205 | 0.8602 | |
| GSE45670 | RHAG | 6005 | 206146_s_at | -0.0456 | 0.6782 | |
| GSE53622 | RHAG | 6005 | 29336 | -0.0140 | 0.9124 | |
| GSE53624 | RHAG | 6005 | 29336 | -0.2718 | 0.0042 | |
| GSE63941 | RHAG | 6005 | 206146_s_at | 0.1529 | 0.3396 | |
| GSE77861 | RHAG | 6005 | 206146_s_at | -0.0400 | 0.8117 | |
| TCGA | RHAG | 6005 | RNAseq | -3.9884 | 0.0041 |
Upregulated datasets: 0; Downregulated datasets: 1.
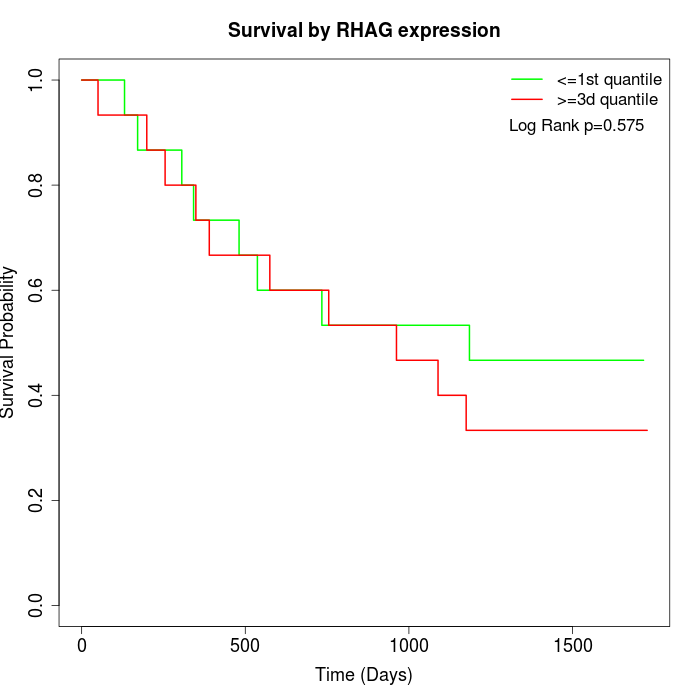
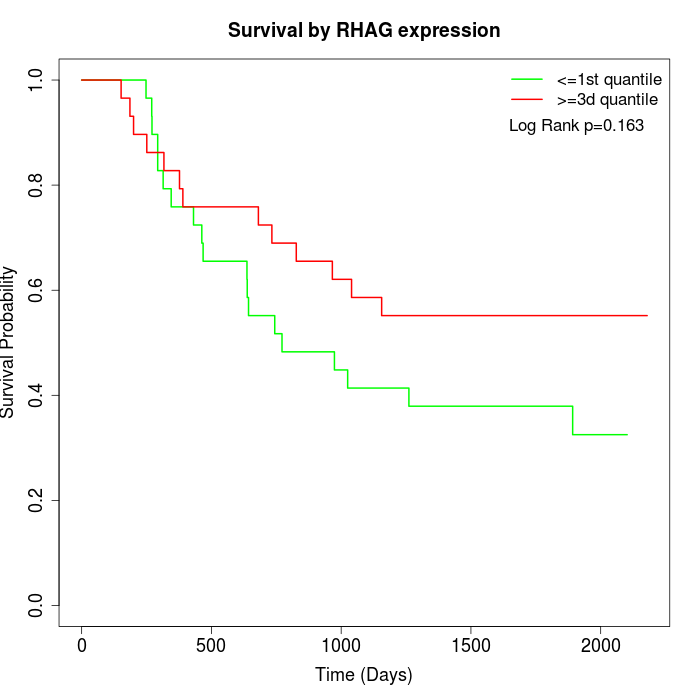
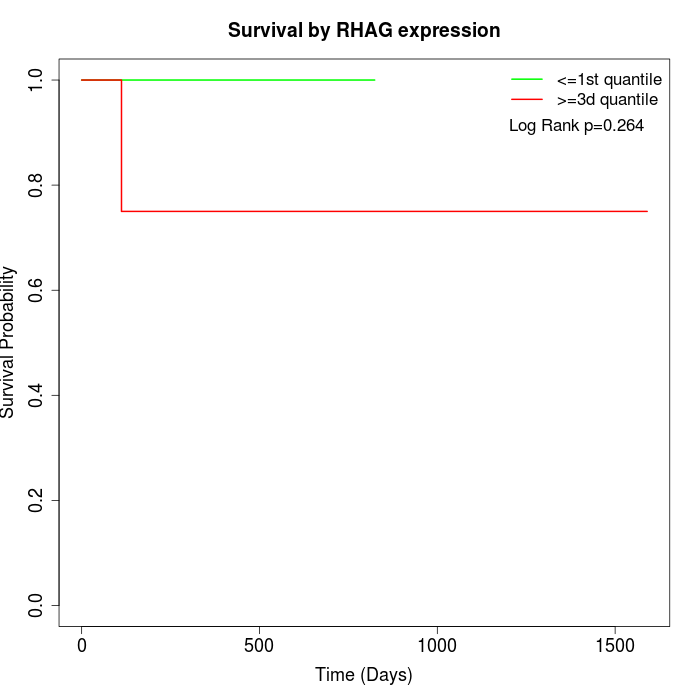
Survival by RHAG expression:
Note: Click image to view full size file.
Copy number change of RHAG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RHAG | 6005 | 5 | 2 | 23 | |
| GSE20123 | RHAG | 6005 | 5 | 2 | 23 | |
| GSE43470 | RHAG | 6005 | 5 | 0 | 38 | |
| GSE46452 | RHAG | 6005 | 2 | 10 | 47 | |
| GSE47630 | RHAG | 6005 | 7 | 5 | 28 | |
| GSE54993 | RHAG | 6005 | 3 | 1 | 66 | |
| GSE54994 | RHAG | 6005 | 8 | 4 | 41 | |
| GSE60625 | RHAG | 6005 | 0 | 3 | 8 | |
| GSE74703 | RHAG | 6005 | 5 | 0 | 31 | |
| GSE74704 | RHAG | 6005 | 2 | 1 | 17 | |
| TCGA | RHAG | 6005 | 21 | 12 | 63 |
Total number of gains: 63; Total number of losses: 40; Total Number of normals: 385.
Somatic mutations of RHAG:
Generating mutation plots.
Highly correlated genes for RHAG:
Showing top 20/1071 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RHAG | DPF3 | 0.772508 | 3 | 0 | 3 |
| RHAG | SSTR4 | 0.758781 | 3 | 0 | 3 |
| RHAG | SNAP25 | 0.757578 | 3 | 0 | 3 |
| RHAG | ATRNL1 | 0.752018 | 3 | 0 | 3 |
| RHAG | RASL10B | 0.750673 | 3 | 0 | 3 |
| RHAG | PHLDB1 | 0.747426 | 3 | 0 | 3 |
| RHAG | CADM3 | 0.746457 | 3 | 0 | 3 |
| RHAG | GDF3 | 0.739731 | 3 | 0 | 3 |
| RHAG | GDF5 | 0.73881 | 3 | 0 | 3 |
| RHAG | MAPK4 | 0.737796 | 4 | 0 | 4 |
| RHAG | MAS1 | 0.736057 | 3 | 0 | 3 |
| RHAG | GGN | 0.732169 | 3 | 0 | 3 |
| RHAG | MTAP | 0.731663 | 4 | 0 | 4 |
| RHAG | EDDM3A | 0.731074 | 3 | 0 | 3 |
| RHAG | CACNA1F | 0.731059 | 4 | 0 | 3 |
| RHAG | TAS2R3 | 0.730354 | 3 | 0 | 3 |
| RHAG | LGALS14 | 0.729825 | 3 | 0 | 3 |
| RHAG | LAMC3 | 0.72382 | 3 | 0 | 3 |
| RHAG | CACNB1 | 0.717404 | 3 | 0 | 3 |
| RHAG | CAPN11 | 0.716688 | 3 | 0 | 3 |
For details and further investigation, click here