| Full name: nuclear receptor subfamily 0 group B member 2 | Alias Symbol: SHP | ||
| Type: protein-coding gene | Cytoband: 1p36.11 | ||
| Entrez ID: 8431 | HGNC ID: HGNC:7961 | Ensembl Gene: ENSG00000131910 | OMIM ID: 604630 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NR0B2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04976 | Bile secretion |
Expression of NR0B2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NR0B2 | 8431 | 206410_at | 0.0664 | 0.8106 | |
| GSE20347 | NR0B2 | 8431 | 206410_at | -0.0479 | 0.6143 | |
| GSE23400 | NR0B2 | 8431 | 206410_at | -0.0960 | 0.0427 | |
| GSE26886 | NR0B2 | 8431 | 206410_at | 0.0892 | 0.3123 | |
| GSE29001 | NR0B2 | 8431 | 206410_at | -0.1467 | 0.4385 | |
| GSE38129 | NR0B2 | 8431 | 206410_at | -0.2214 | 0.1841 | |
| GSE45670 | NR0B2 | 8431 | 206410_at | 0.1071 | 0.2197 | |
| GSE53622 | NR0B2 | 8431 | 45084 | -0.2463 | 0.2297 | |
| GSE53624 | NR0B2 | 8431 | 45084 | -0.4722 | 0.0119 | |
| GSE63941 | NR0B2 | 8431 | 206410_at | -0.0161 | 0.9167 | |
| GSE77861 | NR0B2 | 8431 | 206410_at | -0.1422 | 0.2912 | |
| TCGA | NR0B2 | 8431 | RNAseq | -6.2287 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
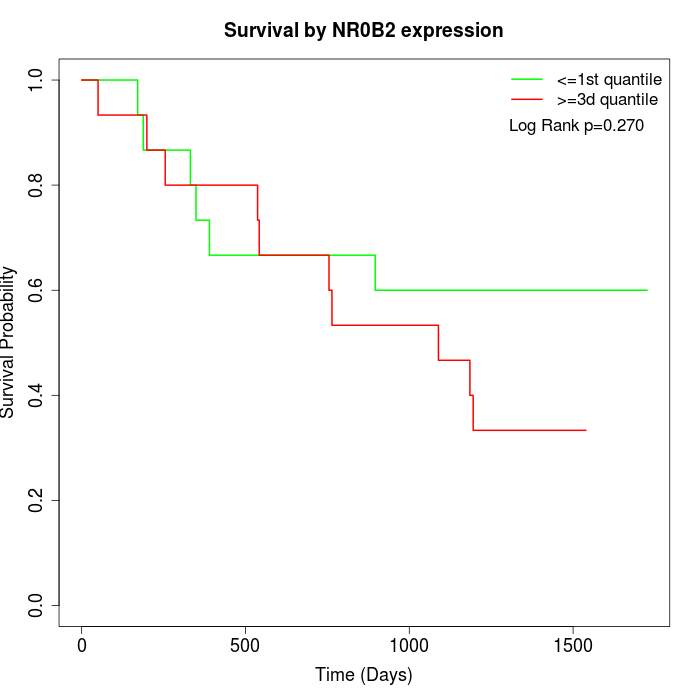
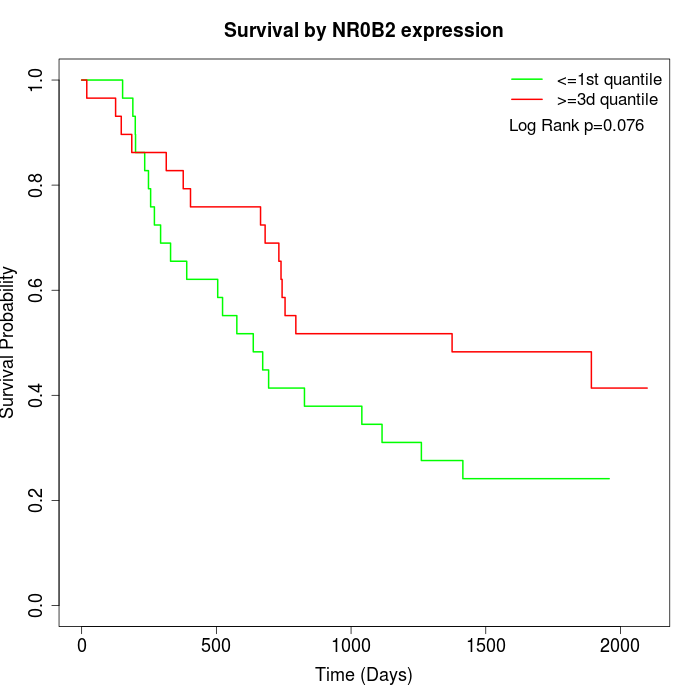
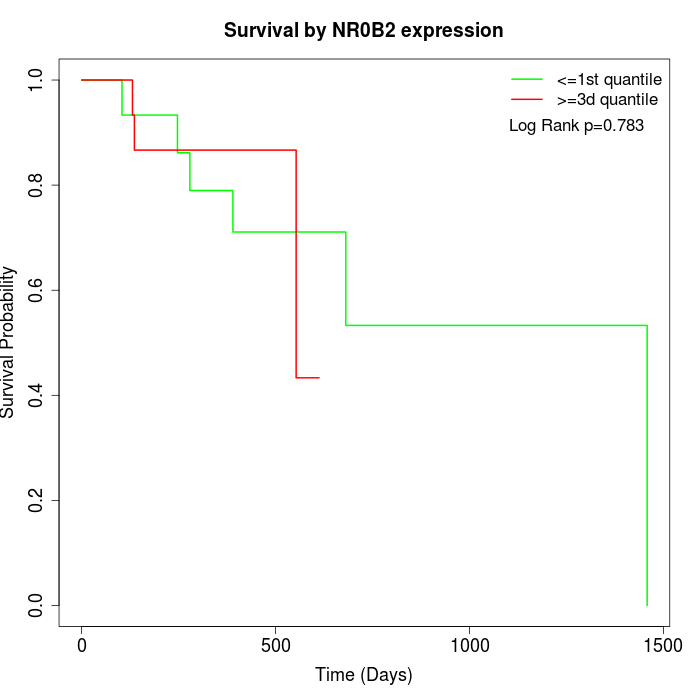
Survival by NR0B2 expression:
Note: Click image to view full size file.
Copy number change of NR0B2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NR0B2 | 8431 | 0 | 6 | 24 | |
| GSE20123 | NR0B2 | 8431 | 0 | 5 | 25 | |
| GSE43470 | NR0B2 | 8431 | 2 | 6 | 35 | |
| GSE46452 | NR0B2 | 8431 | 5 | 1 | 53 | |
| GSE47630 | NR0B2 | 8431 | 8 | 3 | 29 | |
| GSE54993 | NR0B2 | 8431 | 2 | 1 | 67 | |
| GSE54994 | NR0B2 | 8431 | 10 | 4 | 39 | |
| GSE60625 | NR0B2 | 8431 | 0 | 0 | 11 | |
| GSE74703 | NR0B2 | 8431 | 1 | 4 | 31 | |
| GSE74704 | NR0B2 | 8431 | 0 | 0 | 20 | |
| TCGA | NR0B2 | 8431 | 10 | 24 | 62 |
Total number of gains: 38; Total number of losses: 54; Total Number of normals: 396.
Somatic mutations of NR0B2:
Generating mutation plots.
Highly correlated genes for NR0B2:
Showing top 20/999 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NR0B2 | KCNE2 | 0.730848 | 5 | 0 | 5 |
| NR0B2 | PGC | 0.725577 | 7 | 0 | 7 |
| NR0B2 | IFNA4 | 0.708044 | 4 | 0 | 4 |
| NR0B2 | ZNF696 | 0.705709 | 3 | 0 | 3 |
| NR0B2 | RNF17 | 0.705633 | 3 | 0 | 3 |
| NR0B2 | KCNJ10 | 0.699151 | 4 | 0 | 4 |
| NR0B2 | KCNA10 | 0.696426 | 3 | 0 | 3 |
| NR0B2 | TPSG1 | 0.694966 | 5 | 0 | 5 |
| NR0B2 | MBL2 | 0.694124 | 3 | 0 | 3 |
| NR0B2 | CYP11B2 | 0.687731 | 3 | 0 | 3 |
| NR0B2 | ZNF358 | 0.686275 | 3 | 0 | 3 |
| NR0B2 | SMIM6 | 0.682653 | 4 | 0 | 3 |
| NR0B2 | IL9 | 0.679746 | 3 | 0 | 3 |
| NR0B2 | OR11A1 | 0.678298 | 3 | 0 | 3 |
| NR0B2 | DPEP1 | 0.675795 | 4 | 0 | 4 |
| NR0B2 | GSN-AS1 | 0.675674 | 3 | 0 | 3 |
| NR0B2 | FGF4 | 0.673251 | 4 | 0 | 4 |
| NR0B2 | LIPF | 0.672519 | 6 | 0 | 5 |
| NR0B2 | DLGAP2-AS1 | 0.67046 | 4 | 0 | 4 |
| NR0B2 | ATP4A | 0.669615 | 6 | 0 | 6 |
For details and further investigation, click here