| Full name: fibrinogen like 1 | Alias Symbol: HFREP-1 | ||
| Type: protein-coding gene | Cytoband: 8p22 | ||
| Entrez ID: 2267 | HGNC ID: HGNC:3695 | Ensembl Gene: ENSG00000104760 | OMIM ID: 605776 |
| Drug and gene relationship at DGIdb | |||
Expression of FGL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FGL1 | 2267 | 205305_at | 0.1245 | 0.6149 | |
| GSE20347 | FGL1 | 2267 | 205305_at | 0.0571 | 0.3915 | |
| GSE23400 | FGL1 | 2267 | 205305_at | -0.0555 | 0.2390 | |
| GSE26886 | FGL1 | 2267 | 205305_at | 0.2211 | 0.0518 | |
| GSE29001 | FGL1 | 2267 | 205305_at | 0.0675 | 0.7330 | |
| GSE38129 | FGL1 | 2267 | 205305_at | -0.0147 | 0.8964 | |
| GSE45670 | FGL1 | 2267 | 205305_at | 0.0404 | 0.6646 | |
| GSE53622 | FGL1 | 2267 | 11701 | -0.1052 | 0.1375 | |
| GSE53624 | FGL1 | 2267 | 11701 | -0.0368 | 0.6470 | |
| GSE63941 | FGL1 | 2267 | 205305_at | 0.1747 | 0.5912 | |
| GSE77861 | FGL1 | 2267 | 205305_at | 0.0902 | 0.4386 | |
| GSE97050 | FGL1 | 2267 | A_23_P20484 | -0.1439 | 0.4509 | |
| TCGA | FGL1 | 2267 | RNAseq | -0.5284 | 0.6998 |
Upregulated datasets: 0; Downregulated datasets: 0.
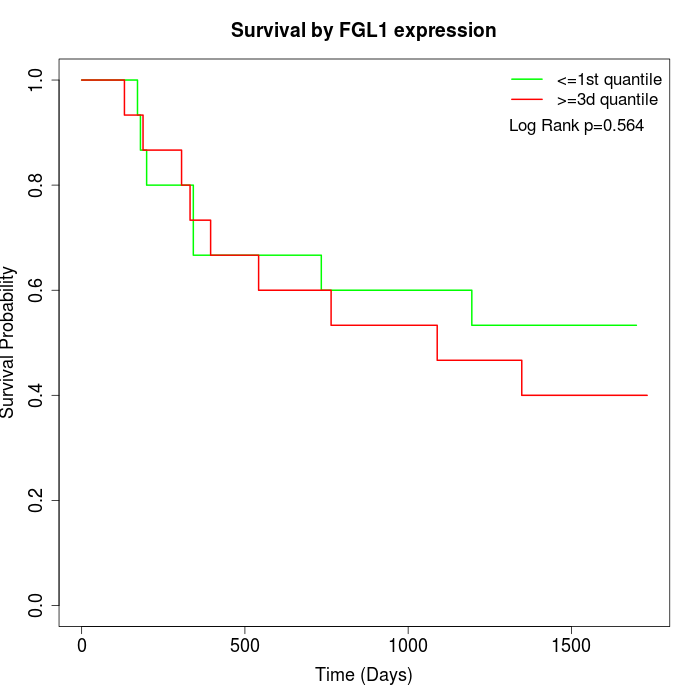
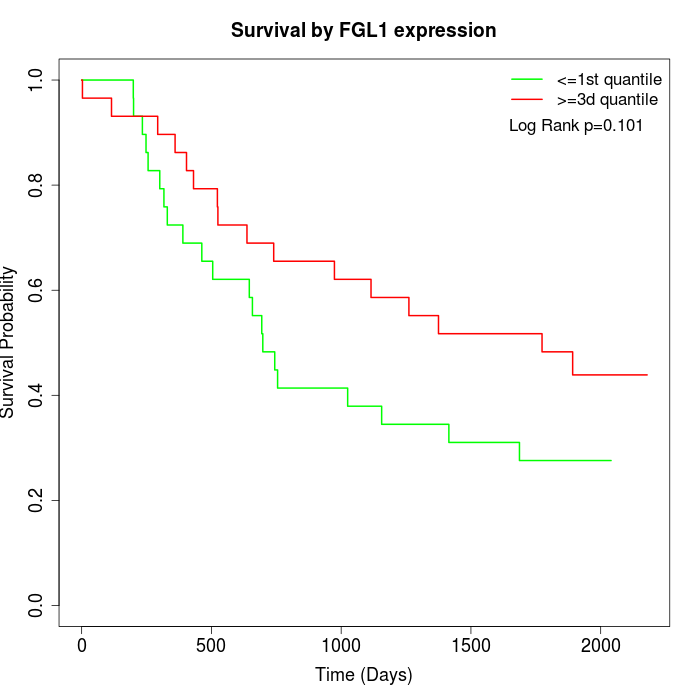
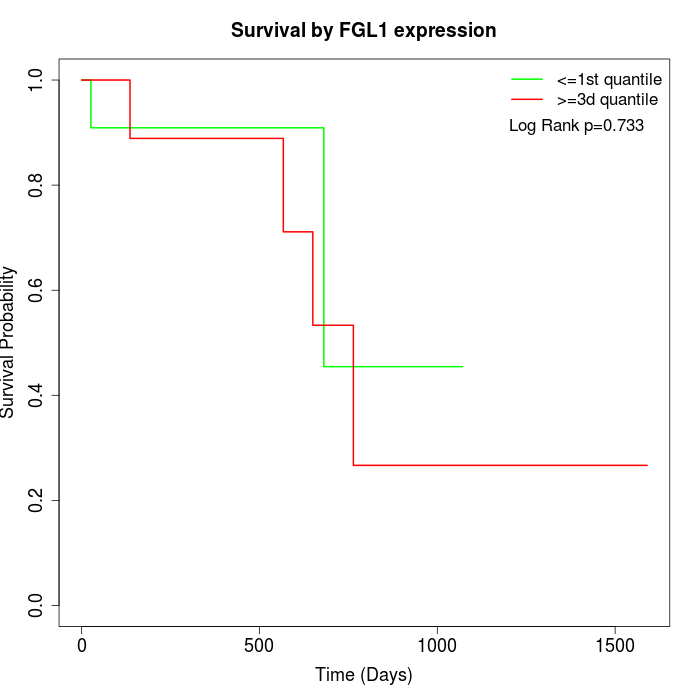
Survival by FGL1 expression:
Note: Click image to view full size file.
Copy number change of FGL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FGL1 | 2267 | 3 | 10 | 17 | |
| GSE20123 | FGL1 | 2267 | 4 | 10 | 16 | |
| GSE43470 | FGL1 | 2267 | 4 | 8 | 31 | |
| GSE46452 | FGL1 | 2267 | 14 | 13 | 32 | |
| GSE47630 | FGL1 | 2267 | 10 | 8 | 22 | |
| GSE54993 | FGL1 | 2267 | 2 | 14 | 54 | |
| GSE54994 | FGL1 | 2267 | 8 | 18 | 27 | |
| GSE60625 | FGL1 | 2267 | 3 | 0 | 8 | |
| GSE74703 | FGL1 | 2267 | 4 | 7 | 25 | |
| GSE74704 | FGL1 | 2267 | 3 | 7 | 10 | |
| TCGA | FGL1 | 2267 | 14 | 41 | 41 |
Total number of gains: 69; Total number of losses: 136; Total Number of normals: 283.
Somatic mutations of FGL1:
Generating mutation plots.
Highly correlated genes for FGL1:
Showing top 20/360 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FGL1 | DNAJA4 | 0.726608 | 4 | 0 | 4 |
| FGL1 | BRS3 | 0.69805 | 3 | 0 | 3 |
| FGL1 | ZMAT4 | 0.697085 | 3 | 0 | 3 |
| FGL1 | TNPO1 | 0.694781 | 3 | 0 | 3 |
| FGL1 | GHSR | 0.689458 | 4 | 0 | 3 |
| FGL1 | RNF185-AS1 | 0.680749 | 4 | 0 | 3 |
| FGL1 | IFNA17 | 0.674059 | 4 | 0 | 4 |
| FGL1 | DAB1 | 0.671139 | 4 | 0 | 4 |
| FGL1 | MYOD1 | 0.658467 | 5 | 0 | 4 |
| FGL1 | ZPBP | 0.657327 | 3 | 0 | 3 |
| FGL1 | GALNT6 | 0.649574 | 3 | 0 | 3 |
| FGL1 | CRX | 0.64463 | 3 | 0 | 3 |
| FGL1 | CBX8 | 0.639443 | 3 | 0 | 3 |
| FGL1 | KCNA4 | 0.637437 | 4 | 0 | 4 |
| FGL1 | STMN2 | 0.63734 | 4 | 0 | 3 |
| FGL1 | NTNG1 | 0.63726 | 4 | 0 | 4 |
| FGL1 | WAS | 0.635297 | 3 | 0 | 3 |
| FGL1 | AMELY | 0.634988 | 4 | 0 | 4 |
| FGL1 | ZNF483 | 0.632038 | 3 | 0 | 3 |
| FGL1 | CETN1 | 0.627525 | 4 | 0 | 3 |
For details and further investigation, click here