| Full name: potassium voltage-gated channel subfamily A member 4 | Alias Symbol: Kv1.4|HK1|HPCN2 | ||
| Type: protein-coding gene | Cytoband: 11p14.1 | ||
| Entrez ID: 3739 | HGNC ID: HGNC:6222 | Ensembl Gene: ENSG00000182255 | OMIM ID: 176266 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNA4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNA4 | 3739 | 207248_at | 0.1895 | 0.3366 | |
| GSE20347 | KCNA4 | 3739 | 207248_at | 0.0127 | 0.8696 | |
| GSE23400 | KCNA4 | 3739 | 207248_at | -0.1131 | 0.0009 | |
| GSE26886 | KCNA4 | 3739 | 207248_at | -0.1420 | 0.1903 | |
| GSE29001 | KCNA4 | 3739 | 207248_at | -0.0173 | 0.9346 | |
| GSE38129 | KCNA4 | 3739 | 207248_at | -0.0663 | 0.3010 | |
| GSE45670 | KCNA4 | 3739 | 207248_at | -0.1184 | 0.0923 | |
| GSE53622 | KCNA4 | 3739 | 30545 | -0.2128 | 0.2284 | |
| GSE53624 | KCNA4 | 3739 | 30545 | 0.0581 | 0.4872 | |
| GSE63941 | KCNA4 | 3739 | 207248_at | 0.0460 | 0.7287 | |
| GSE77861 | KCNA4 | 3739 | 207248_at | -0.0834 | 0.2917 | |
| TCGA | KCNA4 | 3739 | RNAseq | -1.2534 | 0.3251 |
Upregulated datasets: 0; Downregulated datasets: 0.
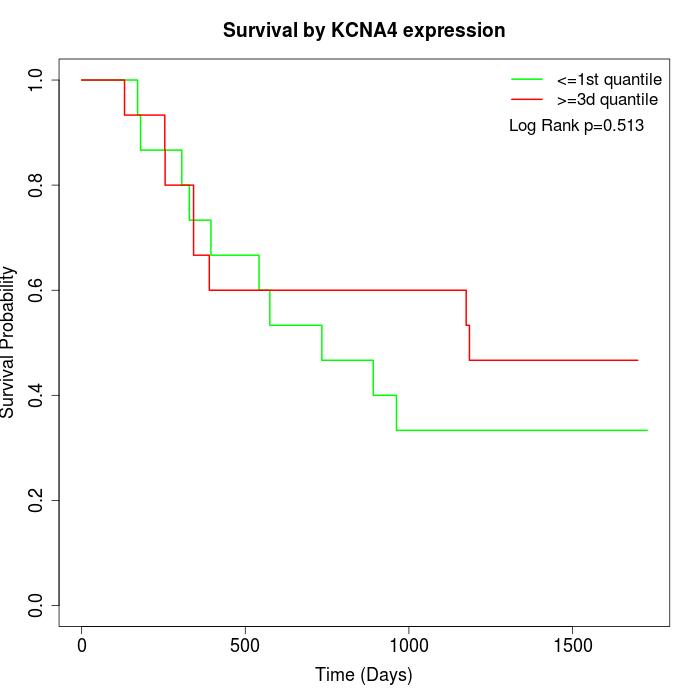
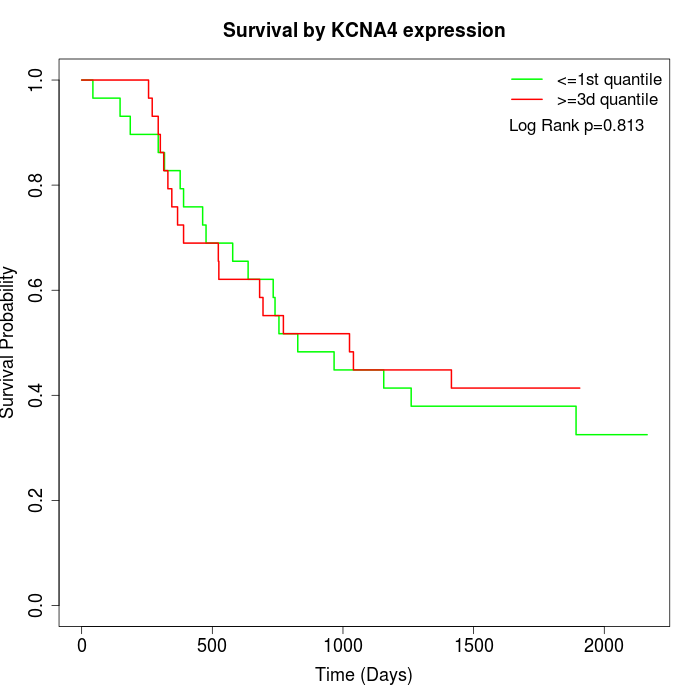
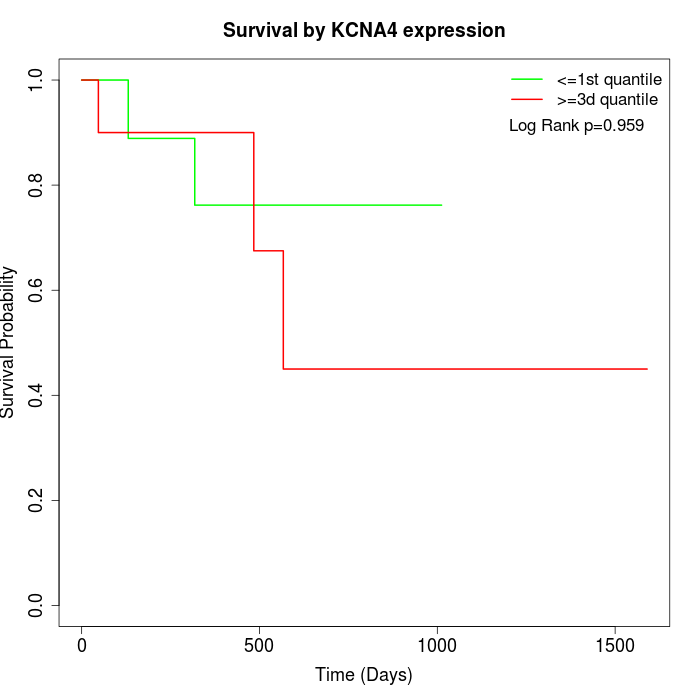
Survival by KCNA4 expression:
Note: Click image to view full size file.
Copy number change of KCNA4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNA4 | 3739 | 1 | 9 | 20 | |
| GSE20123 | KCNA4 | 3739 | 1 | 8 | 21 | |
| GSE43470 | KCNA4 | 3739 | 1 | 4 | 38 | |
| GSE46452 | KCNA4 | 3739 | 8 | 5 | 46 | |
| GSE47630 | KCNA4 | 3739 | 3 | 10 | 27 | |
| GSE54993 | KCNA4 | 3739 | 3 | 0 | 67 | |
| GSE54994 | KCNA4 | 3739 | 5 | 10 | 38 | |
| GSE60625 | KCNA4 | 3739 | 0 | 0 | 11 | |
| GSE74703 | KCNA4 | 3739 | 1 | 2 | 33 | |
| GSE74704 | KCNA4 | 3739 | 0 | 8 | 12 | |
| TCGA | KCNA4 | 3739 | 12 | 25 | 59 |
Total number of gains: 35; Total number of losses: 81; Total Number of normals: 372.
Somatic mutations of KCNA4:
Generating mutation plots.
Highly correlated genes for KCNA4:
Showing top 20/768 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNA4 | SLC9A7 | 0.777321 | 3 | 0 | 3 |
| KCNA4 | GCG | 0.725394 | 3 | 0 | 3 |
| KCNA4 | ADCYAP1 | 0.709076 | 3 | 0 | 3 |
| KCNA4 | PHF7 | 0.705859 | 6 | 0 | 6 |
| KCNA4 | PRL | 0.705403 | 4 | 0 | 4 |
| KCNA4 | LUZP4 | 0.70184 | 5 | 0 | 5 |
| KCNA4 | KCNF1 | 0.698515 | 4 | 0 | 4 |
| KCNA4 | SLC17A1 | 0.697481 | 4 | 0 | 4 |
| KCNA4 | P2RX3 | 0.694821 | 3 | 0 | 3 |
| KCNA4 | RARA | 0.694301 | 3 | 0 | 3 |
| KCNA4 | PDE6B | 0.692553 | 3 | 0 | 3 |
| KCNA4 | ARX | 0.690321 | 3 | 0 | 3 |
| KCNA4 | EDDM3A | 0.684822 | 3 | 0 | 3 |
| KCNA4 | PAPOLB | 0.684388 | 3 | 0 | 3 |
| KCNA4 | CCDC30 | 0.6776 | 5 | 0 | 5 |
| KCNA4 | POU4F3 | 0.675717 | 3 | 0 | 3 |
| KCNA4 | UBE2D4 | 0.673443 | 3 | 0 | 3 |
| KCNA4 | GPR26 | 0.672027 | 3 | 0 | 3 |
| KCNA4 | NTNG1 | 0.671788 | 3 | 0 | 3 |
| KCNA4 | IFNA17 | 0.669681 | 6 | 0 | 6 |
For details and further investigation, click here