| Full name: FGR proto-oncogene, Src family tyrosine kinase | Alias Symbol: c-fgr|p55c-fgr | ||
| Type: protein-coding gene | Cytoband: 1p35.3 | ||
| Entrez ID: 2268 | HGNC ID: HGNC:3697 | Ensembl Gene: ENSG00000000938 | OMIM ID: 164940 |
| Drug and gene relationship at DGIdb | |||
FGR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa05169 | Epstein-Barr virus infection |
Expression of FGR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FGR | 2268 | 208438_s_at | 0.3471 | 0.6175 | |
| GSE20347 | FGR | 2268 | 208438_s_at | -0.0796 | 0.5214 | |
| GSE23400 | FGR | 2268 | 208438_s_at | -0.0951 | 0.0683 | |
| GSE26886 | FGR | 2268 | 208438_s_at | -0.3833 | 0.1147 | |
| GSE29001 | FGR | 2268 | 208438_s_at | -0.2858 | 0.1369 | |
| GSE38129 | FGR | 2268 | 208438_s_at | -0.0800 | 0.5653 | |
| GSE45670 | FGR | 2268 | 208438_s_at | -0.4146 | 0.0536 | |
| GSE53622 | FGR | 2268 | 49515 | 0.1547 | 0.1274 | |
| GSE53624 | FGR | 2268 | 49515 | 0.1218 | 0.1148 | |
| GSE63941 | FGR | 2268 | 208438_s_at | -0.1453 | 0.4064 | |
| GSE77861 | FGR | 2268 | 208438_s_at | -0.2190 | 0.0296 | |
| GSE97050 | FGR | 2268 | A_23_P103932 | 0.3731 | 0.1518 | |
| SRP007169 | FGR | 2268 | RNAseq | 1.4444 | 0.0940 | |
| SRP008496 | FGR | 2268 | RNAseq | 0.0732 | 0.8393 | |
| SRP064894 | FGR | 2268 | RNAseq | 0.7495 | 0.0339 | |
| SRP133303 | FGR | 2268 | RNAseq | 0.0930 | 0.6712 | |
| SRP159526 | FGR | 2268 | RNAseq | -0.5143 | 0.2882 | |
| SRP193095 | FGR | 2268 | RNAseq | 0.6213 | 0.0082 | |
| SRP219564 | FGR | 2268 | RNAseq | 0.0848 | 0.7726 | |
| TCGA | FGR | 2268 | RNAseq | -0.0095 | 0.9452 |
Upregulated datasets: 0; Downregulated datasets: 0.
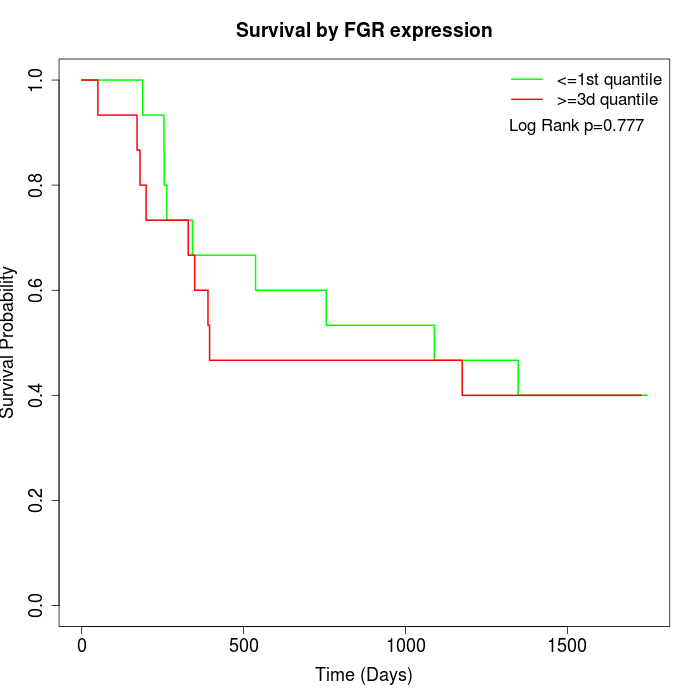
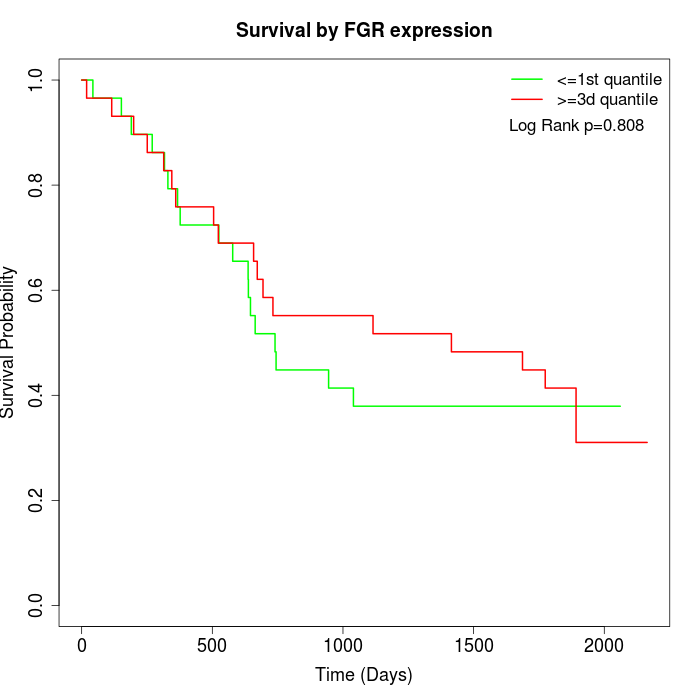
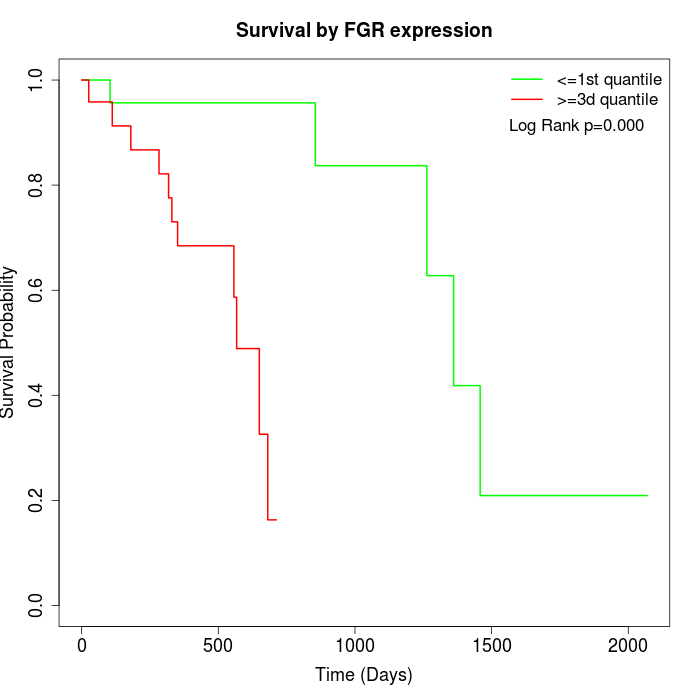
Survival by FGR expression:
Note: Click image to view full size file.
Copy number change of FGR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FGR | 2268 | 0 | 6 | 24 | |
| GSE20123 | FGR | 2268 | 0 | 5 | 25 | |
| GSE43470 | FGR | 2268 | 3 | 6 | 34 | |
| GSE46452 | FGR | 2268 | 5 | 1 | 53 | |
| GSE47630 | FGR | 2268 | 8 | 3 | 29 | |
| GSE54993 | FGR | 2268 | 2 | 1 | 67 | |
| GSE54994 | FGR | 2268 | 10 | 4 | 39 | |
| GSE60625 | FGR | 2268 | 0 | 0 | 11 | |
| GSE74703 | FGR | 2268 | 2 | 4 | 30 | |
| GSE74704 | FGR | 2268 | 0 | 0 | 20 | |
| TCGA | FGR | 2268 | 11 | 23 | 62 |
Total number of gains: 41; Total number of losses: 53; Total Number of normals: 394.
Somatic mutations of FGR:
Generating mutation plots.
Highly correlated genes for FGR:
Showing top 20/248 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FGR | BRINP2 | 0.701068 | 3 | 0 | 3 |
| FGR | KIR3DL1 | 0.686393 | 4 | 0 | 4 |
| FGR | FERMT3 | 0.685028 | 4 | 0 | 3 |
| FGR | GABRB2 | 0.685004 | 3 | 0 | 3 |
| FGR | AKNA | 0.676898 | 4 | 0 | 3 |
| FGR | TRIM31 | 0.673758 | 3 | 0 | 3 |
| FGR | TDRD1 | 0.672419 | 3 | 0 | 3 |
| FGR | TUSC1 | 0.667292 | 3 | 0 | 3 |
| FGR | HK3 | 0.667213 | 4 | 0 | 3 |
| FGR | ELSPBP1 | 0.663222 | 4 | 0 | 3 |
| FGR | CSHL1 | 0.662785 | 4 | 0 | 4 |
| FGR | ITGA4 | 0.66137 | 4 | 0 | 4 |
| FGR | RGSL1 | 0.660315 | 3 | 0 | 3 |
| FGR | ORM1 | 0.659936 | 3 | 0 | 3 |
| FGR | FAM78A | 0.656295 | 3 | 0 | 3 |
| FGR | SLA | 0.655481 | 10 | 0 | 9 |
| FGR | KIR3DL3 | 0.651038 | 5 | 0 | 5 |
| FGR | MMP19 | 0.646458 | 5 | 0 | 4 |
| FGR | CD300LF | 0.64511 | 7 | 0 | 6 |
| FGR | BTNL8 | 0.643799 | 4 | 0 | 4 |
For details and further investigation, click here