| Full name: BMP/retinoic acid inducible neural specific 2 | Alias Symbol: DBCCR1L2 | ||
| Type: protein-coding gene | Cytoband: 1q25.2 | ||
| Entrez ID: 57795 | HGNC ID: HGNC:13746 | Ensembl Gene: ENSG00000198797 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of BRINP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BRINP2 | 57795 | 214822_at | 0.0338 | 0.9044 | |
| GSE20347 | BRINP2 | 57795 | 214822_at | -0.0152 | 0.8671 | |
| GSE23400 | BRINP2 | 57795 | 214822_at | -0.2432 | 0.0000 | |
| GSE26886 | BRINP2 | 57795 | 214822_at | -0.0863 | 0.3563 | |
| GSE29001 | BRINP2 | 57795 | 214822_at | -0.3712 | 0.0233 | |
| GSE38129 | BRINP2 | 57795 | 214822_at | -0.0641 | 0.4850 | |
| GSE45670 | BRINP2 | 57795 | 214822_at | 0.0154 | 0.8956 | |
| GSE53622 | BRINP2 | 57795 | 30267 | 0.2067 | 0.0939 | |
| GSE53624 | BRINP2 | 57795 | 30267 | 0.6253 | 0.0000 | |
| GSE63941 | BRINP2 | 57795 | 214822_at | -0.0913 | 0.4962 | |
| GSE77861 | BRINP2 | 57795 | 214822_at | -0.1790 | 0.1498 |
Upregulated datasets: 0; Downregulated datasets: 0.
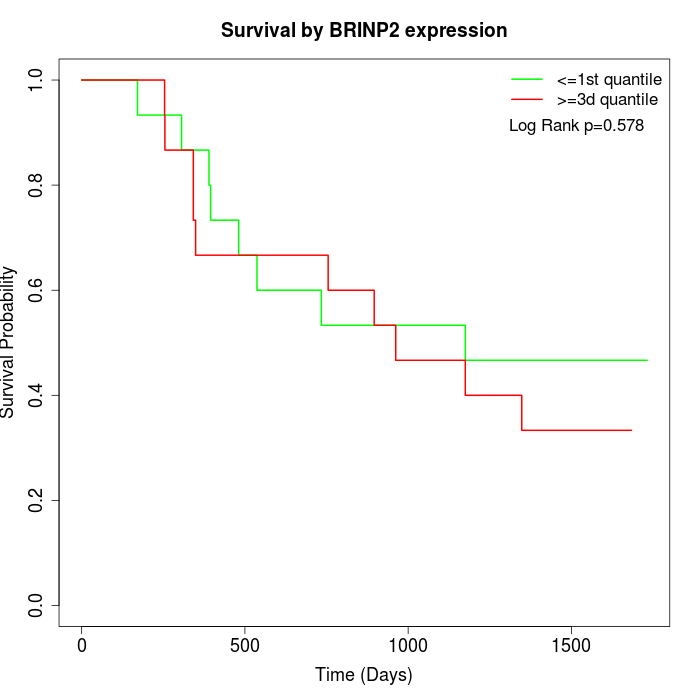
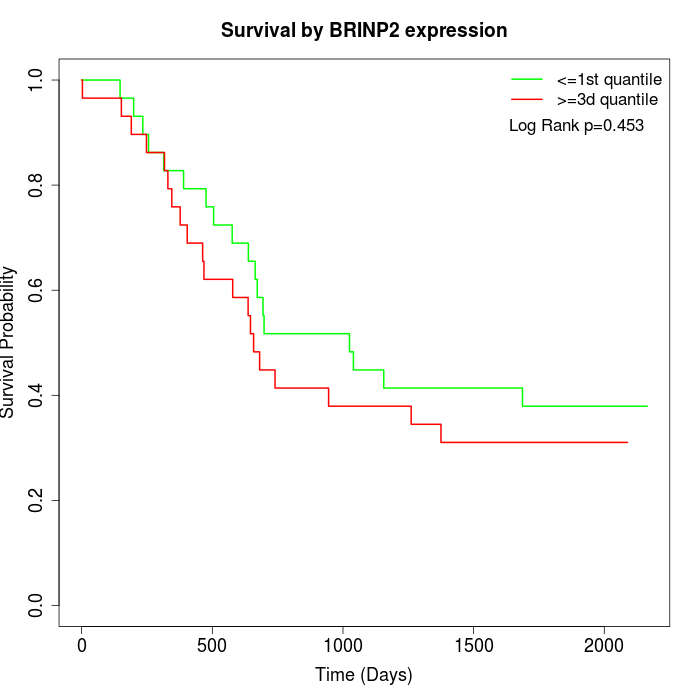
Survival by BRINP2 expression:
Note: Click image to view full size file.
Copy number change of BRINP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BRINP2 | 57795 | 12 | 0 | 18 | |
| GSE20123 | BRINP2 | 57795 | 12 | 0 | 18 | |
| GSE43470 | BRINP2 | 57795 | 7 | 1 | 35 | |
| GSE46452 | BRINP2 | 57795 | 3 | 1 | 55 | |
| GSE47630 | BRINP2 | 57795 | 14 | 0 | 26 | |
| GSE54993 | BRINP2 | 57795 | 0 | 6 | 64 | |
| GSE54994 | BRINP2 | 57795 | 15 | 0 | 38 | |
| GSE60625 | BRINP2 | 57795 | 0 | 0 | 11 | |
| GSE74703 | BRINP2 | 57795 | 7 | 1 | 28 | |
| GSE74704 | BRINP2 | 57795 | 5 | 0 | 15 |
Total number of gains: 75; Total number of losses: 9; Total Number of normals: 308.
Somatic mutations of BRINP2:
Generating mutation plots.
Highly correlated genes for BRINP2:
Showing top 20/928 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BRINP2 | PRPH2 | 0.759423 | 3 | 0 | 3 |
| BRINP2 | HRC | 0.75106 | 3 | 0 | 3 |
| BRINP2 | NCR2 | 0.741183 | 4 | 0 | 4 |
| BRINP2 | KIAA1614 | 0.738884 | 4 | 0 | 4 |
| BRINP2 | CER1 | 0.737554 | 5 | 0 | 5 |
| BRINP2 | CCDC7 | 0.73468 | 4 | 0 | 4 |
| BRINP2 | CLCNKB | 0.732692 | 3 | 0 | 3 |
| BRINP2 | SIGLEC5 | 0.728018 | 4 | 0 | 4 |
| BRINP2 | ADAM11 | 0.724877 | 4 | 0 | 4 |
| BRINP2 | IHH | 0.720526 | 4 | 0 | 4 |
| BRINP2 | ACR | 0.719866 | 4 | 0 | 4 |
| BRINP2 | ABCA4 | 0.713094 | 3 | 0 | 3 |
| BRINP2 | MMP25 | 0.710808 | 4 | 0 | 3 |
| BRINP2 | GDF5 | 0.710415 | 4 | 0 | 4 |
| BRINP2 | APBA1 | 0.709395 | 4 | 0 | 4 |
| BRINP2 | TUB | 0.707848 | 4 | 0 | 4 |
| BRINP2 | CYP11B2 | 0.707108 | 3 | 0 | 3 |
| BRINP2 | OR2H2 | 0.704456 | 4 | 0 | 3 |
| BRINP2 | GLP1R | 0.703624 | 4 | 0 | 4 |
| BRINP2 | RGR | 0.703222 | 4 | 0 | 4 |
For details and further investigation, click here