| Full name: epididymal sperm binding protein 1 | Alias Symbol: HE12|E12|EDDM12 | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 64100 | HGNC ID: HGNC:14417 | Ensembl Gene: | OMIM ID: 607443 |
| Drug and gene relationship at DGIdb | |||
Expression of ELSPBP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ELSPBP1 | 64100 | 220366_at | -0.1555 | 0.4014 | |
| GSE20347 | ELSPBP1 | 64100 | 220366_at | -0.0197 | 0.8399 | |
| GSE23400 | ELSPBP1 | 64100 | 220366_at | -0.1688 | 0.0000 | |
| GSE26886 | ELSPBP1 | 64100 | 220366_at | -0.2185 | 0.1231 | |
| GSE29001 | ELSPBP1 | 64100 | 220366_at | -0.1355 | 0.4108 | |
| GSE38129 | ELSPBP1 | 64100 | 220366_at | -0.1748 | 0.0546 | |
| GSE45670 | ELSPBP1 | 64100 | 220366_at | 0.1160 | 0.1774 | |
| GSE63941 | ELSPBP1 | 64100 | 220366_at | 0.0509 | 0.7620 | |
| GSE77861 | ELSPBP1 | 64100 | 220366_at | -0.2440 | 0.0601 |
Upregulated datasets: 0; Downregulated datasets: 0.
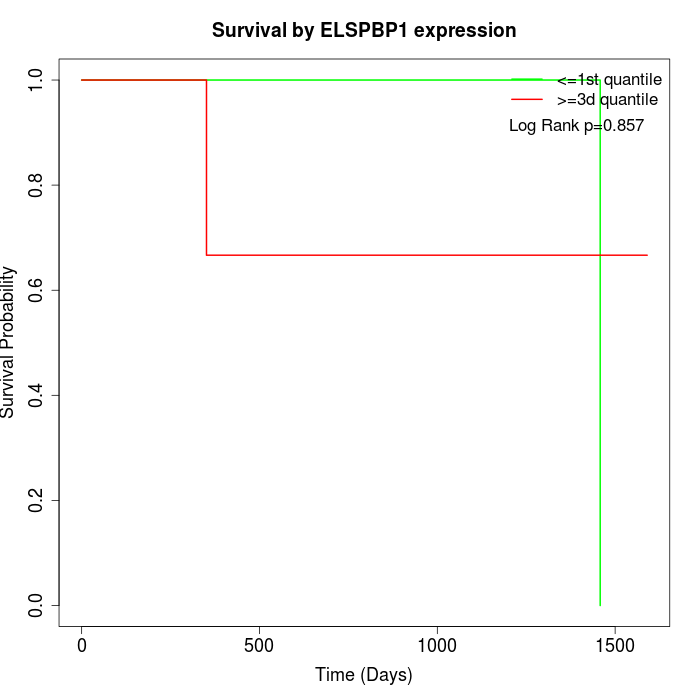
Survival by ELSPBP1 expression:
Note: Click image to view full size file.
Copy number change of ELSPBP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ELSPBP1 | 64100 | 4 | 4 | 22 | |
| GSE20123 | ELSPBP1 | 64100 | 4 | 3 | 23 | |
| GSE43470 | ELSPBP1 | 64100 | 3 | 10 | 30 | |
| GSE46452 | ELSPBP1 | 64100 | 45 | 1 | 13 | |
| GSE47630 | ELSPBP1 | 64100 | 8 | 7 | 25 | |
| GSE54993 | ELSPBP1 | 64100 | 17 | 4 | 49 | |
| GSE54994 | ELSPBP1 | 64100 | 4 | 14 | 35 | |
| GSE60625 | ELSPBP1 | 64100 | 9 | 0 | 2 | |
| GSE74703 | ELSPBP1 | 64100 | 3 | 7 | 26 | |
| GSE74704 | ELSPBP1 | 64100 | 4 | 1 | 15 | |
| TCGA | ELSPBP1 | 64100 | 15 | 19 | 62 |
Total number of gains: 116; Total number of losses: 70; Total Number of normals: 302.
Somatic mutations of ELSPBP1:
Generating mutation plots.
Highly correlated genes for ELSPBP1:
Showing top 20/852 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ELSPBP1 | JAK3 | 0.746807 | 4 | 0 | 4 |
| ELSPBP1 | KHDRBS2 | 0.722931 | 4 | 0 | 4 |
| ELSPBP1 | CHRNA6 | 0.722368 | 3 | 0 | 3 |
| ELSPBP1 | IFNA17 | 0.701414 | 5 | 0 | 5 |
| ELSPBP1 | KIR2DS3 | 0.700227 | 4 | 0 | 4 |
| ELSPBP1 | PROP1 | 0.699407 | 4 | 0 | 4 |
| ELSPBP1 | ART1 | 0.696334 | 5 | 0 | 5 |
| ELSPBP1 | E2F4 | 0.693983 | 6 | 0 | 6 |
| ELSPBP1 | MTAP | 0.693544 | 5 | 0 | 5 |
| ELSPBP1 | AOC2 | 0.690956 | 3 | 0 | 3 |
| ELSPBP1 | ASB4 | 0.688818 | 4 | 0 | 4 |
| ELSPBP1 | ADARB2-AS1 | 0.685983 | 3 | 0 | 3 |
| ELSPBP1 | AKAP6 | 0.685076 | 5 | 0 | 5 |
| ELSPBP1 | PRL | 0.683822 | 4 | 0 | 4 |
| ELSPBP1 | ECE2 | 0.682766 | 4 | 0 | 4 |
| ELSPBP1 | OR1A1 | 0.67549 | 5 | 0 | 4 |
| ELSPBP1 | PAX3 | 0.674333 | 4 | 0 | 4 |
| ELSPBP1 | UBE2D4 | 0.674258 | 3 | 0 | 3 |
| ELSPBP1 | HHLA1 | 0.673257 | 5 | 0 | 5 |
| ELSPBP1 | NPHS2 | 0.670429 | 7 | 0 | 6 |
For details and further investigation, click here