| Full name: integrin subunit alpha 4 | Alias Symbol: CD49d | ||
| Type: protein-coding gene | Cytoband: 2q31.3 | ||
| Entrez ID: 3676 | HGNC ID: HGNC:6140 | Ensembl Gene: ENSG00000115232 | OMIM ID: 192975 |
| Drug and gene relationship at DGIdb | |||
ITGA4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of ITGA4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGA4 | 3676 | 205885_s_at | 0.4917 | 0.6839 | |
| GSE20347 | ITGA4 | 3676 | 205885_s_at | 0.0711 | 0.2583 | |
| GSE23400 | ITGA4 | 3676 | 205884_at | 0.1758 | 0.0004 | |
| GSE26886 | ITGA4 | 3676 | 205885_s_at | 0.0765 | 0.6641 | |
| GSE29001 | ITGA4 | 3676 | 205884_at | 0.0083 | 0.9532 | |
| GSE38129 | ITGA4 | 3676 | 205885_s_at | 0.2995 | 0.0099 | |
| GSE45670 | ITGA4 | 3676 | 205884_at | 0.2917 | 0.1952 | |
| GSE53622 | ITGA4 | 3676 | 138550 | 0.3391 | 0.0007 | |
| GSE53624 | ITGA4 | 3676 | 138550 | -0.0397 | 0.7118 | |
| GSE63941 | ITGA4 | 3676 | 205884_at | -3.2929 | 0.0112 | |
| GSE77861 | ITGA4 | 3676 | 205884_at | 0.1591 | 0.3415 | |
| GSE97050 | ITGA4 | 3676 | A_33_P3353816 | 0.8479 | 0.1399 | |
| SRP007169 | ITGA4 | 3676 | RNAseq | 1.7792 | 0.0017 | |
| SRP008496 | ITGA4 | 3676 | RNAseq | 1.6227 | 0.0000 | |
| SRP064894 | ITGA4 | 3676 | RNAseq | 1.0068 | 0.0015 | |
| SRP133303 | ITGA4 | 3676 | RNAseq | 0.4951 | 0.0363 | |
| SRP159526 | ITGA4 | 3676 | RNAseq | 0.0990 | 0.8835 | |
| SRP193095 | ITGA4 | 3676 | RNAseq | 0.2888 | 0.2286 | |
| SRP219564 | ITGA4 | 3676 | RNAseq | 1.5131 | 0.0303 | |
| TCGA | ITGA4 | 3676 | RNAseq | 0.1516 | 0.2617 |
Upregulated datasets: 4; Downregulated datasets: 1.
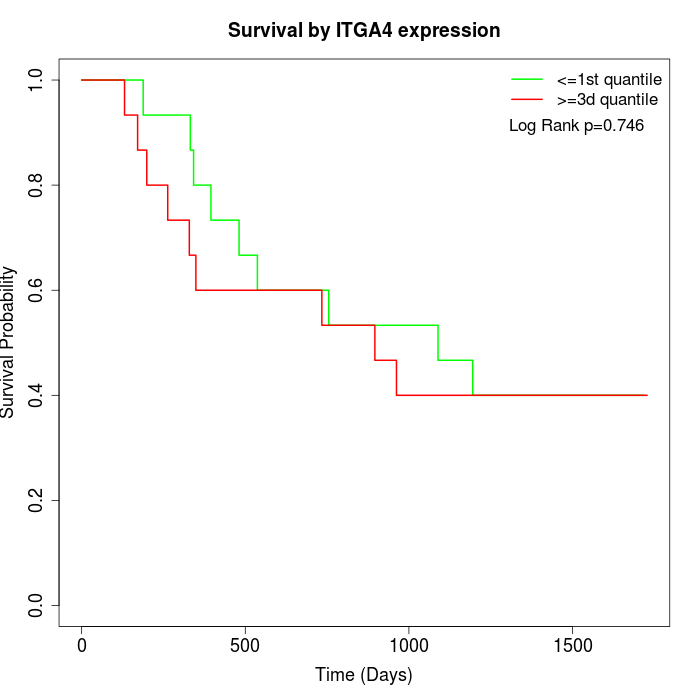
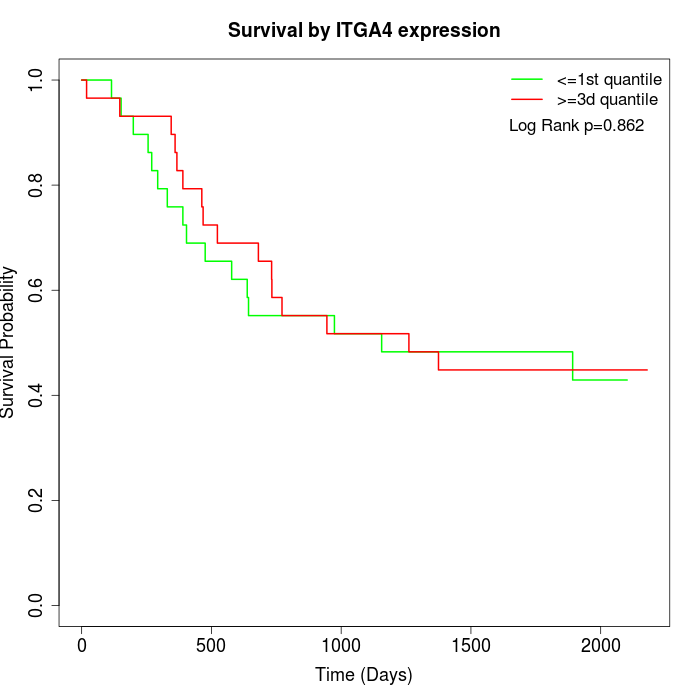
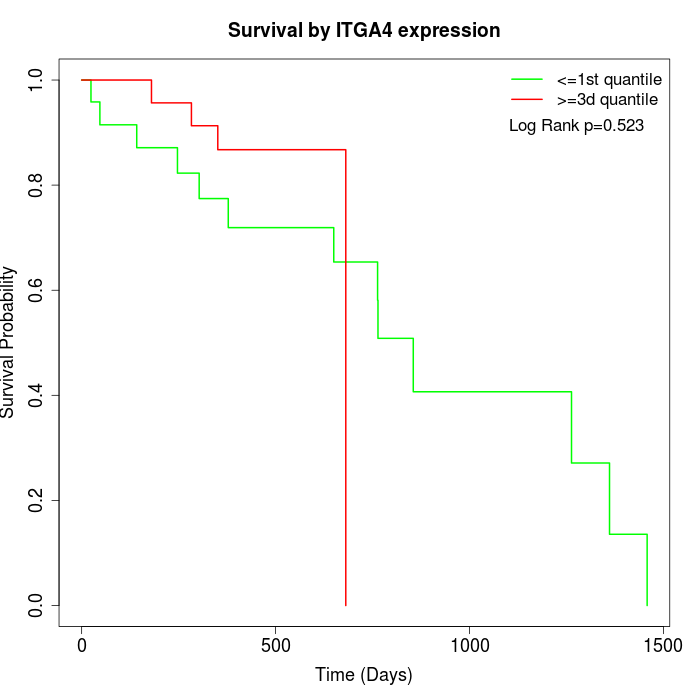
Survival by ITGA4 expression:
Note: Click image to view full size file.
Copy number change of ITGA4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGA4 | 3676 | 5 | 1 | 24 | |
| GSE20123 | ITGA4 | 3676 | 5 | 1 | 24 | |
| GSE43470 | ITGA4 | 3676 | 5 | 1 | 37 | |
| GSE46452 | ITGA4 | 3676 | 1 | 4 | 54 | |
| GSE47630 | ITGA4 | 3676 | 4 | 4 | 32 | |
| GSE54993 | ITGA4 | 3676 | 0 | 5 | 65 | |
| GSE54994 | ITGA4 | 3676 | 12 | 5 | 36 | |
| GSE60625 | ITGA4 | 3676 | 0 | 3 | 8 | |
| GSE74703 | ITGA4 | 3676 | 4 | 1 | 31 | |
| GSE74704 | ITGA4 | 3676 | 2 | 1 | 17 | |
| TCGA | ITGA4 | 3676 | 25 | 8 | 63 |
Total number of gains: 63; Total number of losses: 34; Total Number of normals: 391.
Somatic mutations of ITGA4:
Generating mutation plots.
Highly correlated genes for ITGA4:
Showing top 20/237 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGA4 | FCGR1B | 0.698416 | 5 | 0 | 5 |
| ITGA4 | FAM78A | 0.696058 | 3 | 0 | 3 |
| ITGA4 | CD40 | 0.693947 | 3 | 0 | 3 |
| ITGA4 | ZC3H12D | 0.684228 | 3 | 0 | 3 |
| ITGA4 | ARHGEF6 | 0.675555 | 3 | 0 | 3 |
| ITGA4 | IGDCC4 | 0.670188 | 3 | 0 | 3 |
| ITGA4 | TAGLN2 | 0.661397 | 4 | 0 | 4 |
| ITGA4 | FGR | 0.66137 | 4 | 0 | 4 |
| ITGA4 | SAGE1 | 0.66025 | 3 | 0 | 3 |
| ITGA4 | SERPINE2 | 0.659736 | 3 | 0 | 3 |
| ITGA4 | BNC1 | 0.658863 | 3 | 0 | 3 |
| ITGA4 | COTL1 | 0.652326 | 7 | 0 | 7 |
| ITGA4 | MCOLN2 | 0.650575 | 4 | 0 | 4 |
| ITGA4 | PTGER2 | 0.650124 | 3 | 0 | 3 |
| ITGA4 | AJAP1 | 0.647704 | 3 | 0 | 3 |
| ITGA4 | COL8A1 | 0.646429 | 4 | 0 | 3 |
| ITGA4 | ARHGAP25 | 0.644058 | 5 | 0 | 4 |
| ITGA4 | EXT1 | 0.64348 | 3 | 0 | 3 |
| ITGA4 | IL2RB | 0.643334 | 6 | 0 | 5 |
| ITGA4 | PLXNC1 | 0.638198 | 7 | 0 | 6 |
For details and further investigation, click here