| Full name: fragile histidine triad diadenosine triphosphatase | Alias Symbol: FRA3B|AP3Aase | ||
| Type: protein-coding gene | Cytoband: 3p14.2 | ||
| Entrez ID: 2272 | HGNC ID: HGNC:3701 | Ensembl Gene: ENSG00000189283 | OMIM ID: 601153 |
| Drug and gene relationship at DGIdb | |||
Expression of FHIT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FHIT | 2272 | 206492_at | -0.9018 | 0.0559 | |
| GSE20347 | FHIT | 2272 | 206492_at | -0.6825 | 0.0000 | |
| GSE23400 | FHIT | 2272 | 206492_at | -0.2704 | 0.0000 | |
| GSE26886 | FHIT | 2272 | 206492_at | -0.5585 | 0.0171 | |
| GSE29001 | FHIT | 2272 | 206492_at | -0.6924 | 0.0001 | |
| GSE38129 | FHIT | 2272 | 206492_at | -0.8184 | 0.0000 | |
| GSE45670 | FHIT | 2272 | 206492_at | -0.5589 | 0.0077 | |
| GSE53622 | FHIT | 2272 | 67224 | -0.5838 | 0.0000 | |
| GSE53624 | FHIT | 2272 | 67224 | -0.6553 | 0.0000 | |
| GSE63941 | FHIT | 2272 | 206492_at | 0.5704 | 0.3847 | |
| GSE77861 | FHIT | 2272 | 206492_at | -0.5580 | 0.0017 | |
| GSE97050 | FHIT | 2272 | A_33_P3216601 | -0.2420 | 0.3017 | |
| SRP064894 | FHIT | 2272 | RNAseq | -0.8077 | 0.0001 | |
| SRP133303 | FHIT | 2272 | RNAseq | -0.5941 | 0.0001 | |
| SRP159526 | FHIT | 2272 | RNAseq | -1.2633 | 0.0019 | |
| SRP193095 | FHIT | 2272 | RNAseq | -0.3570 | 0.0337 | |
| SRP219564 | FHIT | 2272 | RNAseq | -0.5479 | 0.3171 | |
| TCGA | FHIT | 2272 | RNAseq | -1.2319 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 2.
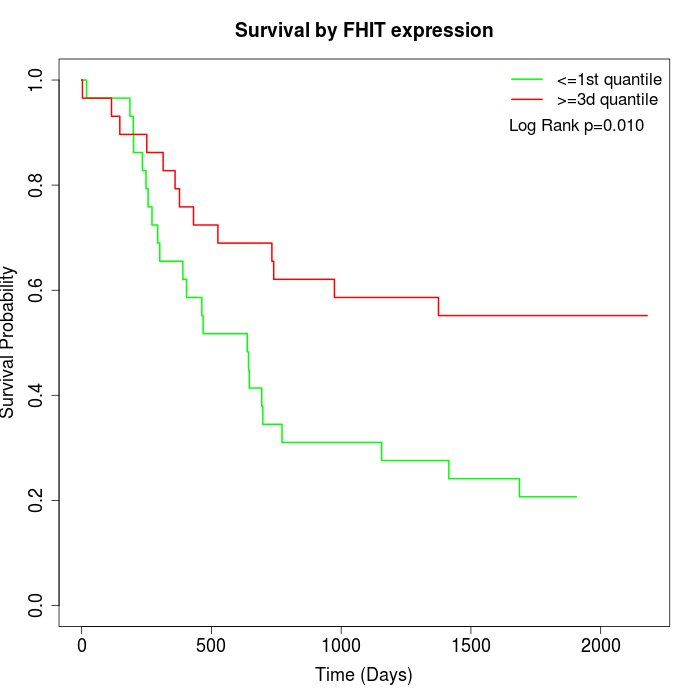
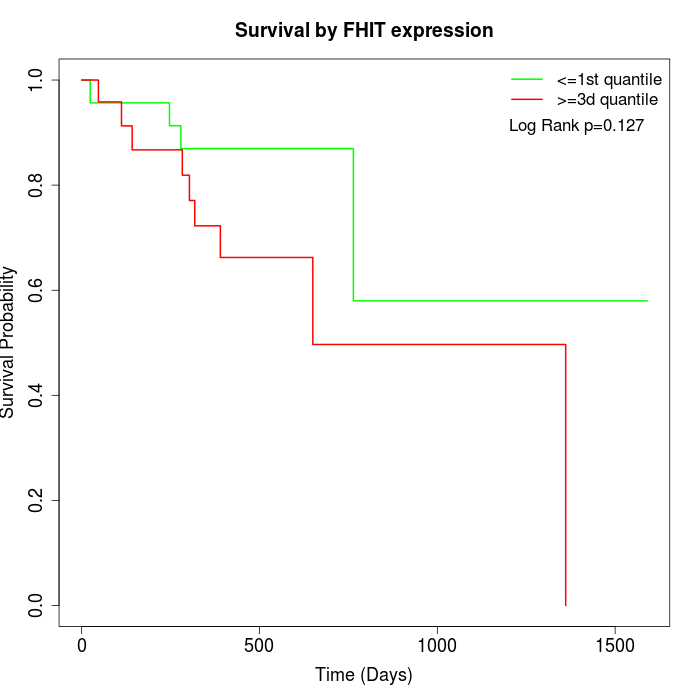
Survival by FHIT expression:
Note: Click image to view full size file.
Copy number change of FHIT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FHIT | 2272 | 0 | 20 | 10 | |
| GSE20123 | FHIT | 2272 | 0 | 20 | 10 | |
| GSE43470 | FHIT | 2272 | 0 | 19 | 24 | |
| GSE46452 | FHIT | 2272 | 2 | 17 | 40 | |
| GSE47630 | FHIT | 2272 | 1 | 24 | 15 | |
| GSE54993 | FHIT | 2272 | 6 | 2 | 62 | |
| GSE54994 | FHIT | 2272 | 0 | 32 | 21 | |
| GSE60625 | FHIT | 2272 | 4 | 1 | 6 | |
| GSE74703 | FHIT | 2272 | 0 | 16 | 20 | |
| GSE74704 | FHIT | 2272 | 0 | 14 | 6 | |
| TCGA | FHIT | 2272 | 1 | 79 | 16 |
Total number of gains: 14; Total number of losses: 244; Total Number of normals: 230.
Somatic mutations of FHIT:
Generating mutation plots.
Highly correlated genes for FHIT:
Showing top 20/542 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FHIT | BSND | 0.763851 | 3 | 0 | 3 |
| FHIT | SCARA5 | 0.750869 | 4 | 0 | 4 |
| FHIT | TMEM217 | 0.733481 | 4 | 0 | 4 |
| FHIT | DUOXA2 | 0.73318 | 3 | 0 | 3 |
| FHIT | NT5C1A | 0.732515 | 4 | 0 | 4 |
| FHIT | RNF186 | 0.729226 | 3 | 0 | 3 |
| FHIT | ANKRD33 | 0.713221 | 4 | 0 | 4 |
| FHIT | OR10G8 | 0.712178 | 3 | 0 | 3 |
| FHIT | B3GNT8 | 0.702394 | 5 | 0 | 3 |
| FHIT | OR51T1 | 0.701257 | 3 | 0 | 3 |
| FHIT | GDF7 | 0.700697 | 3 | 0 | 3 |
| FHIT | TNR | 0.700579 | 5 | 0 | 5 |
| FHIT | RIIAD1 | 0.699177 | 5 | 0 | 5 |
| FHIT | TNXB | 0.696771 | 4 | 0 | 4 |
| FHIT | OR2Z1 | 0.69381 | 3 | 0 | 3 |
| FHIT | EPB41L4A | 0.693722 | 7 | 0 | 7 |
| FHIT | DYRK1B | 0.692385 | 3 | 0 | 3 |
| FHIT | FAM189A2 | 0.691846 | 12 | 0 | 11 |
| FHIT | SHROOM3 | 0.690889 | 6 | 0 | 6 |
| FHIT | ACOX2 | 0.689652 | 9 | 0 | 8 |
For details and further investigation, click here