| Full name: galactose-1-phosphate uridylyltransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 2592 | HGNC ID: HGNC:4135 | Ensembl Gene: ENSG00000213930 | OMIM ID: 606999 |
| Drug and gene relationship at DGIdb | |||
GALT involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04917 | Prolactin signaling pathway |
Expression of GALT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GALT | 2592 | 203179_at | -0.0806 | 0.8455 | |
| GSE20347 | GALT | 2592 | 203179_at | -0.2715 | 0.0175 | |
| GSE23400 | GALT | 2592 | 203179_at | -0.0105 | 0.8920 | |
| GSE26886 | GALT | 2592 | 203179_at | 0.4213 | 0.0200 | |
| GSE29001 | GALT | 2592 | 203179_at | 0.2149 | 0.3548 | |
| GSE38129 | GALT | 2592 | 203179_at | -0.2507 | 0.0102 | |
| GSE45670 | GALT | 2592 | 203179_at | 0.0614 | 0.7616 | |
| GSE53622 | GALT | 2592 | 34460 | -0.1025 | 0.2708 | |
| GSE53624 | GALT | 2592 | 34460 | -0.0288 | 0.7730 | |
| GSE63941 | GALT | 2592 | 203179_at | 0.2759 | 0.5704 | |
| GSE77861 | GALT | 2592 | 203179_at | -0.2652 | 0.1898 | |
| GSE97050 | GALT | 2592 | A_33_P3220415 | -0.1523 | 0.5932 | |
| SRP007169 | GALT | 2592 | RNAseq | 0.0682 | 0.8959 | |
| SRP008496 | GALT | 2592 | RNAseq | -0.0134 | 0.9719 | |
| SRP064894 | GALT | 2592 | RNAseq | 0.4398 | 0.0265 | |
| SRP133303 | GALT | 2592 | RNAseq | -0.6199 | 0.0065 | |
| SRP159526 | GALT | 2592 | RNAseq | -0.5951 | 0.0115 | |
| SRP193095 | GALT | 2592 | RNAseq | -0.1269 | 0.4238 | |
| SRP219564 | GALT | 2592 | RNAseq | 0.1518 | 0.6534 | |
| TCGA | GALT | 2592 | RNAseq | -0.0741 | 0.3018 |
Upregulated datasets: 0; Downregulated datasets: 0.
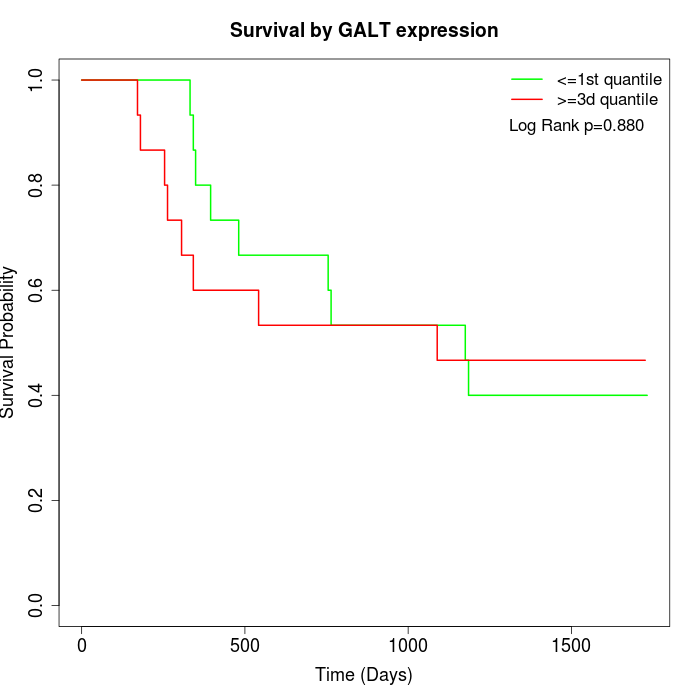
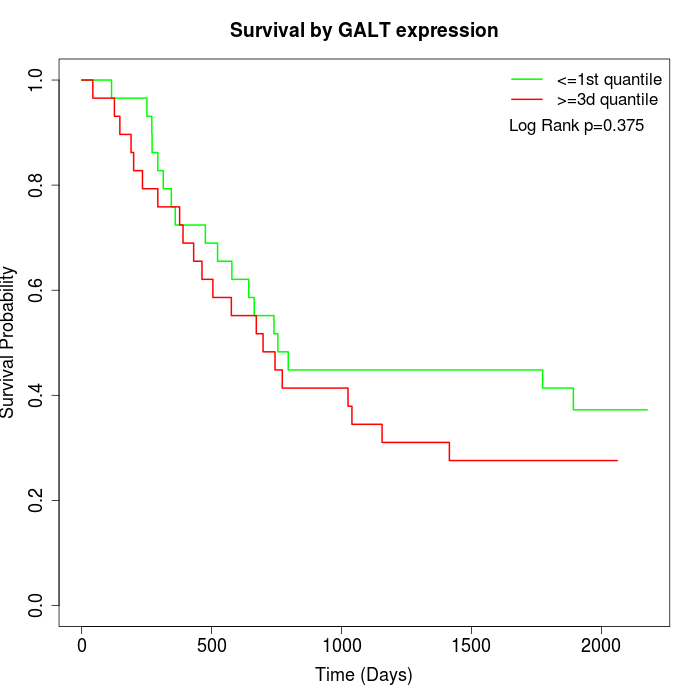
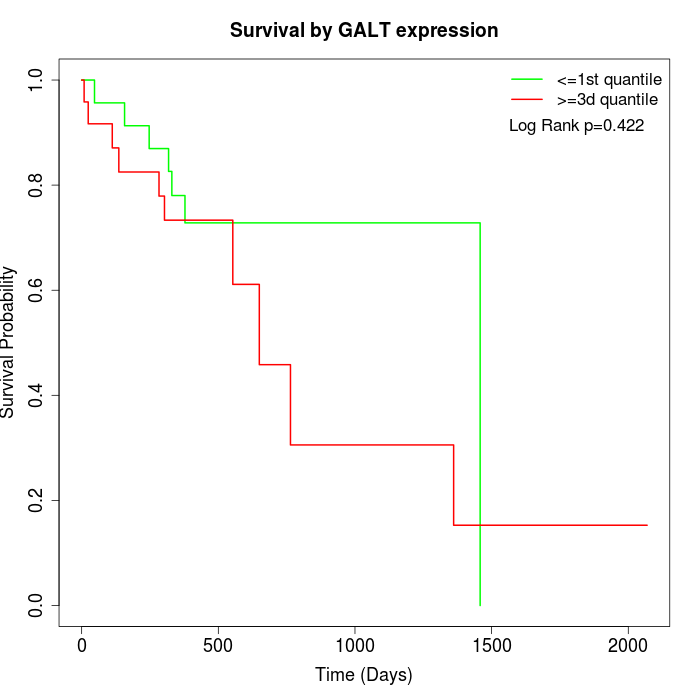
Survival by GALT expression:
Note: Click image to view full size file.
Copy number change of GALT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GALT | 2592 | 4 | 13 | 13 | |
| GSE20123 | GALT | 2592 | 4 | 13 | 13 | |
| GSE43470 | GALT | 2592 | 3 | 10 | 30 | |
| GSE46452 | GALT | 2592 | 6 | 15 | 38 | |
| GSE47630 | GALT | 2592 | 1 | 20 | 19 | |
| GSE54993 | GALT | 2592 | 6 | 0 | 64 | |
| GSE54994 | GALT | 2592 | 6 | 12 | 35 | |
| GSE60625 | GALT | 2592 | 0 | 0 | 11 | |
| GSE74703 | GALT | 2592 | 2 | 7 | 27 | |
| GSE74704 | GALT | 2592 | 0 | 11 | 9 | |
| TCGA | GALT | 2592 | 17 | 44 | 35 |
Total number of gains: 49; Total number of losses: 145; Total Number of normals: 294.
Somatic mutations of GALT:
Generating mutation plots.
Highly correlated genes for GALT:
Showing top 20/30 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GALT | CPA5 | 0.757073 | 3 | 0 | 3 |
| GALT | KCNA7 | 0.705042 | 3 | 0 | 3 |
| GALT | TRIM36 | 0.684262 | 3 | 0 | 3 |
| GALT | ZNF341 | 0.665005 | 3 | 0 | 3 |
| GALT | SIGLEC8 | 0.65474 | 4 | 0 | 4 |
| GALT | ROBO3 | 0.638603 | 4 | 0 | 3 |
| GALT | SHC2 | 0.63832 | 4 | 0 | 3 |
| GALT | MEP1A | 0.633391 | 4 | 0 | 3 |
| GALT | LENG1 | 0.618591 | 4 | 0 | 3 |
| GALT | AZU1 | 0.606648 | 4 | 0 | 3 |
| GALT | CCDC8 | 0.601486 | 4 | 0 | 3 |
| GALT | IGSF21 | 0.590807 | 4 | 0 | 3 |
| GALT | SHARPIN | 0.588665 | 4 | 0 | 3 |
| GALT | ZNF821 | 0.585432 | 4 | 0 | 3 |
| GALT | PHF1 | 0.581152 | 6 | 0 | 3 |
| GALT | OR1F1 | 0.580851 | 4 | 0 | 3 |
| GALT | CYP46A1 | 0.579956 | 3 | 0 | 3 |
| GALT | NKX2-8 | 0.576269 | 4 | 0 | 3 |
| GALT | TMEM101 | 0.572168 | 3 | 0 | 3 |
| GALT | POLR1E | 0.571447 | 5 | 0 | 4 |
For details and further investigation, click here