| Full name: glial fibrillary acidic protein | Alias Symbol: FLJ45472 | ||
| Type: protein-coding gene | Cytoband: 17q21.31 | ||
| Entrez ID: 2670 | HGNC ID: HGNC:4235 | Ensembl Gene: ENSG00000131095 | OMIM ID: 137780 |
| Drug and gene relationship at DGIdb | |||
GFAP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04630 | Jak-STAT signaling pathway |
Expression of GFAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | GFAP | 2670 | 12869 | 0.2421 | 0.0343 | |
| GSE53624 | GFAP | 2670 | 12869 | 0.2207 | 0.0151 | |
| GSE97050 | GFAP | 2670 | A_33_P3335895 | 0.0312 | 0.9004 | |
| SRP133303 | GFAP | 2670 | RNAseq | -0.0174 | 0.9247 | |
| SRP159526 | GFAP | 2670 | RNAseq | 0.1353 | 0.8298 | |
| SRP193095 | GFAP | 2670 | RNAseq | 0.2351 | 0.1719 | |
| SRP219564 | GFAP | 2670 | RNAseq | 0.7796 | 0.1502 | |
| TCGA | GFAP | 2670 | RNAseq | -0.1917 | 0.7493 |
Upregulated datasets: 0; Downregulated datasets: 0.
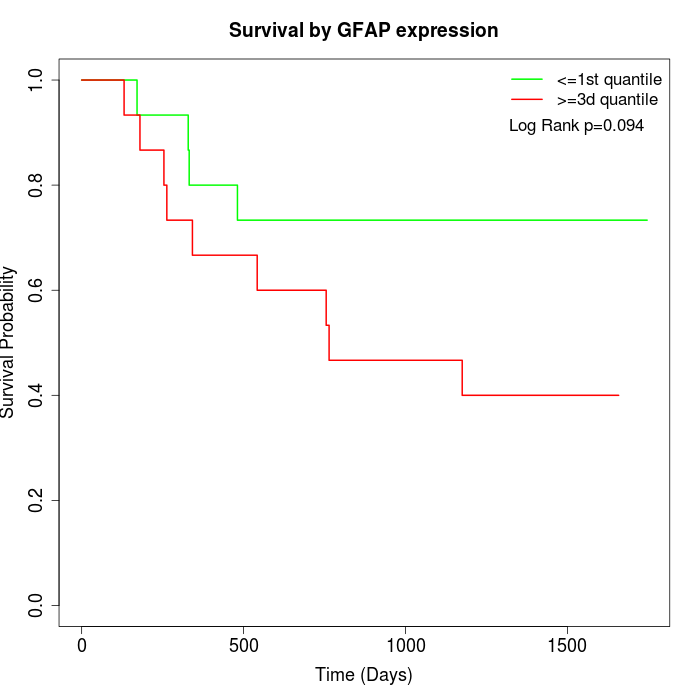
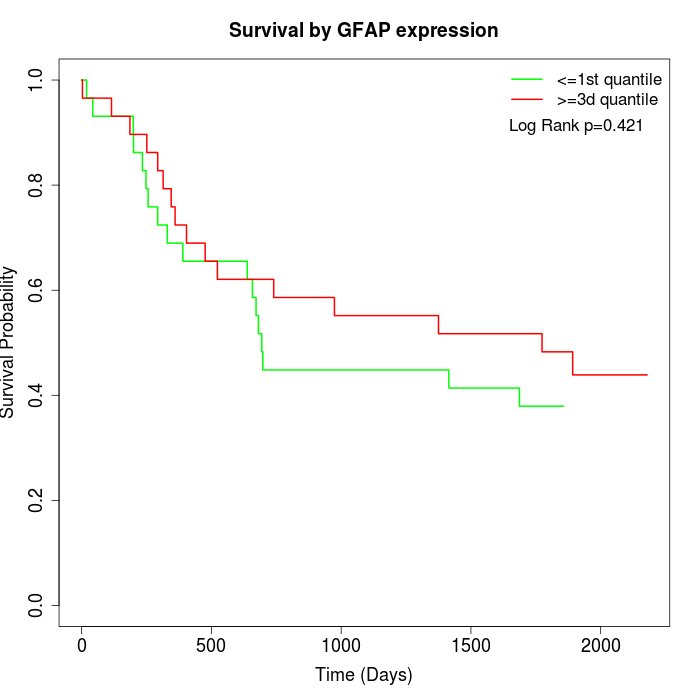
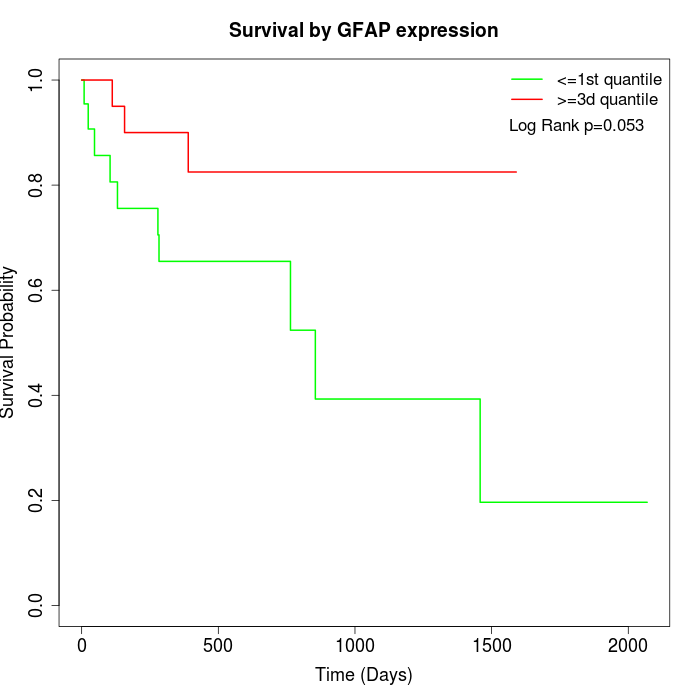
Survival by GFAP expression:
Note: Click image to view full size file.
Copy number change of GFAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GFAP | 2670 | 6 | 2 | 22 | |
| GSE20123 | GFAP | 2670 | 6 | 2 | 22 | |
| GSE43470 | GFAP | 2670 | 1 | 2 | 40 | |
| GSE46452 | GFAP | 2670 | 32 | 0 | 27 | |
| GSE47630 | GFAP | 2670 | 9 | 0 | 31 | |
| GSE54993 | GFAP | 2670 | 3 | 4 | 63 | |
| GSE54994 | GFAP | 2670 | 9 | 6 | 38 | |
| GSE60625 | GFAP | 2670 | 4 | 0 | 7 | |
| GSE74703 | GFAP | 2670 | 1 | 1 | 34 | |
| GSE74704 | GFAP | 2670 | 4 | 1 | 15 | |
| TCGA | GFAP | 2670 | 27 | 6 | 63 |
Total number of gains: 102; Total number of losses: 24; Total Number of normals: 362.
Somatic mutations of GFAP:
Generating mutation plots.
Highly correlated genes for GFAP:
Showing top 20/48 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GFAP | TMEM63B | 0.752959 | 3 | 0 | 3 |
| GFAP | ZNF761 | 0.747077 | 3 | 0 | 3 |
| GFAP | NF2 | 0.740545 | 3 | 0 | 3 |
| GFAP | KRTAP12-2 | 0.707276 | 3 | 0 | 3 |
| GFAP | RBM17 | 0.704944 | 3 | 0 | 3 |
| GFAP | CCDC154 | 0.698863 | 3 | 0 | 3 |
| GFAP | PARP6 | 0.698138 | 3 | 0 | 3 |
| GFAP | KLHL33 | 0.693351 | 3 | 0 | 3 |
| GFAP | COL11A2 | 0.692068 | 3 | 0 | 3 |
| GFAP | OXTR | 0.685811 | 3 | 0 | 3 |
| GFAP | LRRC27 | 0.683325 | 3 | 0 | 3 |
| GFAP | GLCE | 0.681351 | 3 | 0 | 3 |
| GFAP | ANKRD65 | 0.677275 | 3 | 0 | 3 |
| GFAP | SZT2 | 0.665868 | 3 | 0 | 3 |
| GFAP | SLC38A7 | 0.65587 | 3 | 0 | 3 |
| GFAP | GRIA4 | 0.655366 | 3 | 0 | 3 |
| GFAP | OR2T33 | 0.652524 | 3 | 0 | 3 |
| GFAP | C6orf163 | 0.652217 | 3 | 0 | 3 |
| GFAP | NXNL2 | 0.637942 | 3 | 0 | 3 |
| GFAP | SLC26A5 | 0.635999 | 3 | 0 | 3 |
For details and further investigation, click here