| Full name: neurofibromin 2 | Alias Symbol: merlin|ACN|SCH|BANF | ||
| Type: protein-coding gene | Cytoband: 22q12.2 | ||
| Entrez ID: 4771 | HGNC ID: HGNC:7773 | Ensembl Gene: ENSG00000186575 | OMIM ID: 607379 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NF2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway |
Expression of NF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NF2 | 4771 | 218915_at | 0.4758 | 0.2333 | |
| GSE20347 | NF2 | 4771 | 218915_at | -0.3927 | 0.0004 | |
| GSE23400 | NF2 | 4771 | 218915_at | -0.0757 | 0.1205 | |
| GSE26886 | NF2 | 4771 | 218915_at | -0.1528 | 0.3601 | |
| GSE29001 | NF2 | 4771 | 218915_at | -0.2486 | 0.3176 | |
| GSE38129 | NF2 | 4771 | 218915_at | -0.2937 | 0.0003 | |
| GSE45670 | NF2 | 4771 | 218915_at | 0.0911 | 0.7048 | |
| GSE53622 | NF2 | 4771 | 122523 | 0.1372 | 0.0252 | |
| GSE53624 | NF2 | 4771 | 122523 | -0.0923 | 0.2383 | |
| GSE63941 | NF2 | 4771 | 218915_at | -0.5676 | 0.1933 | |
| GSE77861 | NF2 | 4771 | 218915_at | 0.0196 | 0.9267 | |
| GSE97050 | NF2 | 4771 | A_24_P168726 | -0.0496 | 0.8915 | |
| SRP007169 | NF2 | 4771 | RNAseq | -0.4148 | 0.2634 | |
| SRP008496 | NF2 | 4771 | RNAseq | -0.1126 | 0.6878 | |
| SRP064894 | NF2 | 4771 | RNAseq | 0.1371 | 0.3843 | |
| SRP133303 | NF2 | 4771 | RNAseq | -0.0885 | 0.5006 | |
| SRP159526 | NF2 | 4771 | RNAseq | 0.1616 | 0.4812 | |
| SRP193095 | NF2 | 4771 | RNAseq | 0.0274 | 0.7990 | |
| SRP219564 | NF2 | 4771 | RNAseq | 0.1975 | 0.4447 | |
| TCGA | NF2 | 4771 | RNAseq | -0.0439 | 0.3765 |
Upregulated datasets: 0; Downregulated datasets: 0.
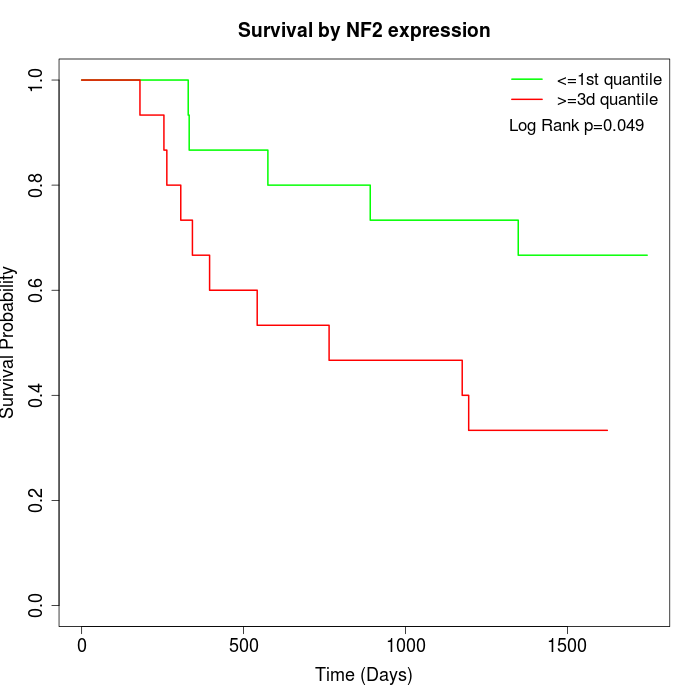
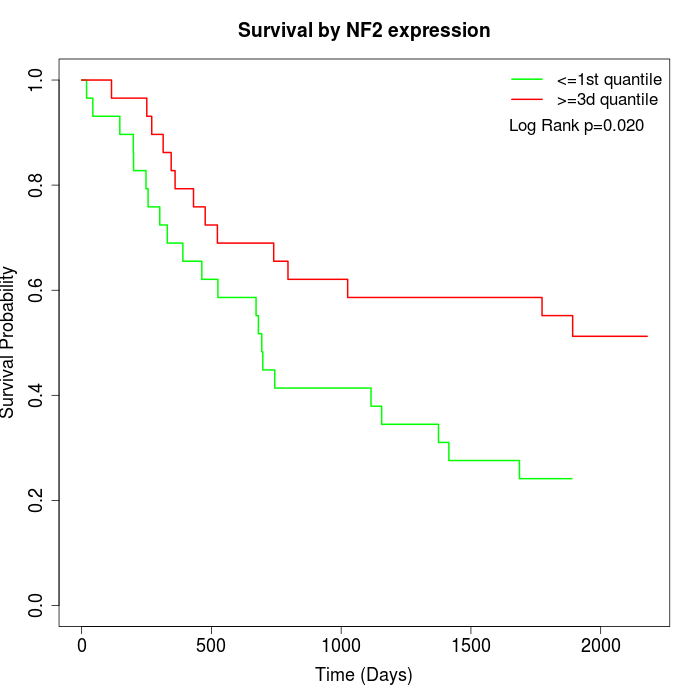
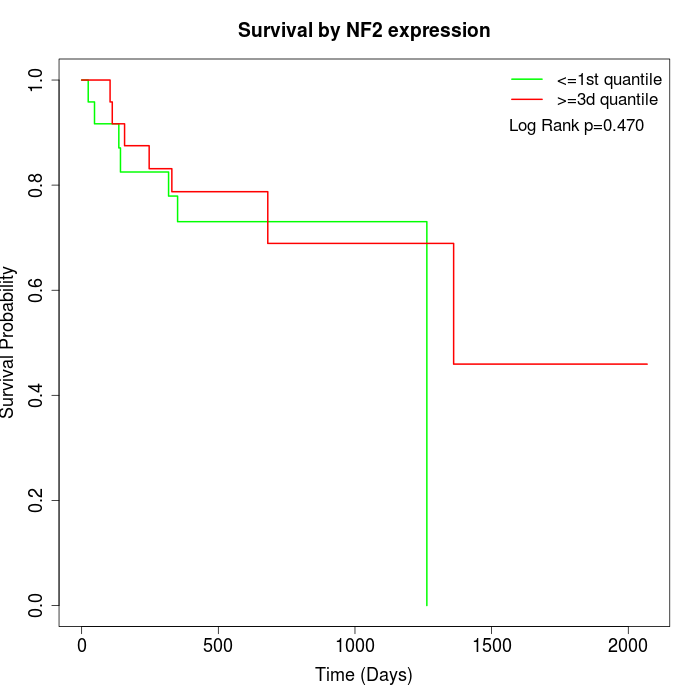
Survival by NF2 expression:
Note: Click image to view full size file.
Copy number change of NF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NF2 | 4771 | 3 | 5 | 22 | |
| GSE20123 | NF2 | 4771 | 3 | 4 | 23 | |
| GSE43470 | NF2 | 4771 | 4 | 5 | 34 | |
| GSE46452 | NF2 | 4771 | 31 | 1 | 27 | |
| GSE47630 | NF2 | 4771 | 8 | 5 | 27 | |
| GSE54993 | NF2 | 4771 | 3 | 6 | 61 | |
| GSE54994 | NF2 | 4771 | 11 | 8 | 34 | |
| GSE60625 | NF2 | 4771 | 5 | 0 | 6 | |
| GSE74703 | NF2 | 4771 | 4 | 4 | 28 | |
| GSE74704 | NF2 | 4771 | 0 | 1 | 19 | |
| TCGA | NF2 | 4771 | 27 | 15 | 54 |
Total number of gains: 99; Total number of losses: 54; Total Number of normals: 335.
Somatic mutations of NF2:
Generating mutation plots.
Highly correlated genes for NF2:
Showing top 20/158 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NF2 | ANKRD65 | 0.822946 | 3 | 0 | 3 |
| NF2 | TATDN2 | 0.806509 | 3 | 0 | 3 |
| NF2 | PREB | 0.775401 | 3 | 0 | 3 |
| NF2 | ZC3H7B | 0.762769 | 3 | 0 | 3 |
| NF2 | STK35 | 0.761985 | 3 | 0 | 3 |
| NF2 | CELA3A | 0.760626 | 3 | 0 | 3 |
| NF2 | SLC26A5 | 0.760377 | 3 | 0 | 3 |
| NF2 | TOR1A | 0.749208 | 3 | 0 | 3 |
| NF2 | GFAP | 0.740545 | 3 | 0 | 3 |
| NF2 | TMEM63B | 0.735286 | 4 | 0 | 3 |
| NF2 | SMAD1 | 0.73375 | 3 | 0 | 3 |
| NF2 | MTCH2 | 0.732546 | 3 | 0 | 3 |
| NF2 | SPEG | 0.723898 | 3 | 0 | 3 |
| NF2 | KIR3DL2 | 0.714485 | 3 | 0 | 3 |
| NF2 | SFTPA2 | 0.71373 | 3 | 0 | 3 |
| NF2 | CYP26C1 | 0.710299 | 3 | 0 | 3 |
| NF2 | ZNF419 | 0.707716 | 3 | 0 | 3 |
| NF2 | BARHL1 | 0.702234 | 3 | 0 | 3 |
| NF2 | ALG11 | 0.701038 | 3 | 0 | 3 |
| NF2 | OR2T33 | 0.698434 | 4 | 0 | 4 |
For details and further investigation, click here