| Full name: golgi associated, gamma adaptin ear containing, ARF binding protein 2 | Alias Symbol: VEAR|KIAA1080 | ||
| Type: protein-coding gene | Cytoband: 16p12.2 | ||
| Entrez ID: 23062 | HGNC ID: HGNC:16064 | Ensembl Gene: ENSG00000103365 | OMIM ID: 606005 |
| Drug and gene relationship at DGIdb | |||
Expression of GGA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GGA2 | 23062 | 213772_s_at | 0.3265 | 0.4943 | |
| GSE20347 | GGA2 | 23062 | 208915_s_at | 0.2425 | 0.0232 | |
| GSE23400 | GGA2 | 23062 | 208915_s_at | 0.2707 | 0.0000 | |
| GSE26886 | GGA2 | 23062 | 208913_at | 0.4598 | 0.1151 | |
| GSE29001 | GGA2 | 23062 | 208913_at | 0.5977 | 0.0550 | |
| GSE38129 | GGA2 | 23062 | 213772_s_at | 0.1966 | 0.0891 | |
| GSE45670 | GGA2 | 23062 | 213772_s_at | 0.1843 | 0.1552 | |
| GSE53622 | GGA2 | 23062 | 2694 | -0.2289 | 0.0035 | |
| GSE53624 | GGA2 | 23062 | 2694 | -0.1757 | 0.0324 | |
| GSE63941 | GGA2 | 23062 | 208915_s_at | 1.7281 | 0.0006 | |
| GSE77861 | GGA2 | 23062 | 208915_s_at | 0.1665 | 0.2385 | |
| GSE97050 | GGA2 | 23062 | A_24_P416257 | 0.0357 | 0.8776 | |
| SRP007169 | GGA2 | 23062 | RNAseq | 0.4507 | 0.1822 | |
| SRP008496 | GGA2 | 23062 | RNAseq | 0.4631 | 0.0268 | |
| SRP064894 | GGA2 | 23062 | RNAseq | -0.0390 | 0.7771 | |
| SRP133303 | GGA2 | 23062 | RNAseq | 0.1175 | 0.4622 | |
| SRP159526 | GGA2 | 23062 | RNAseq | 0.0145 | 0.9557 | |
| SRP193095 | GGA2 | 23062 | RNAseq | 0.1104 | 0.3399 | |
| SRP219564 | GGA2 | 23062 | RNAseq | 0.0028 | 0.9918 | |
| TCGA | GGA2 | 23062 | RNAseq | -0.1459 | 0.0030 |
Upregulated datasets: 1; Downregulated datasets: 0.
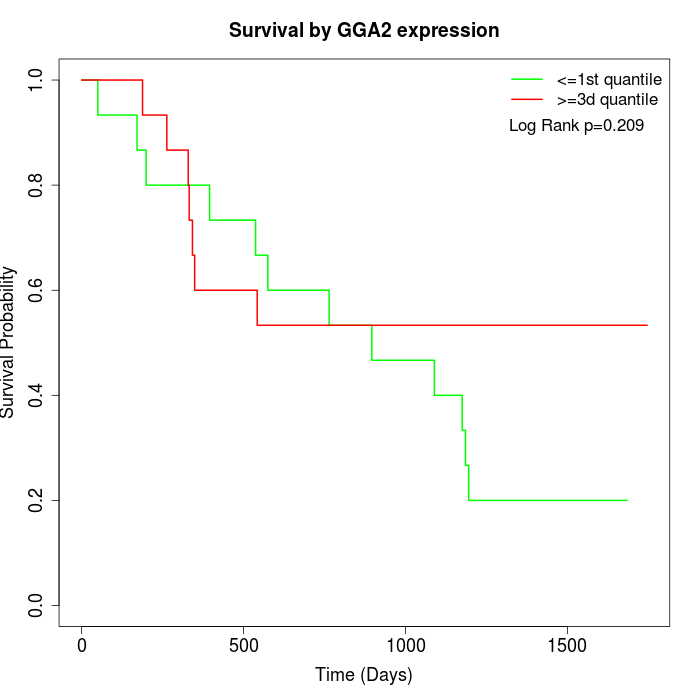
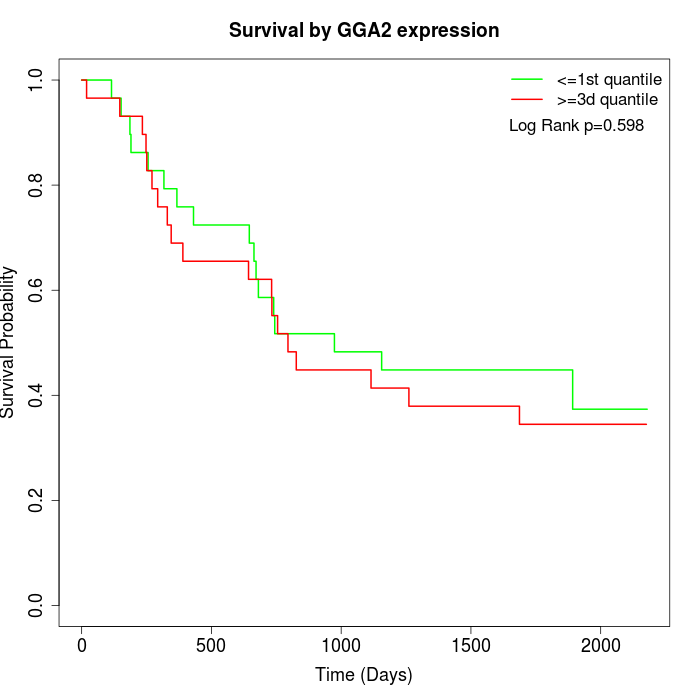
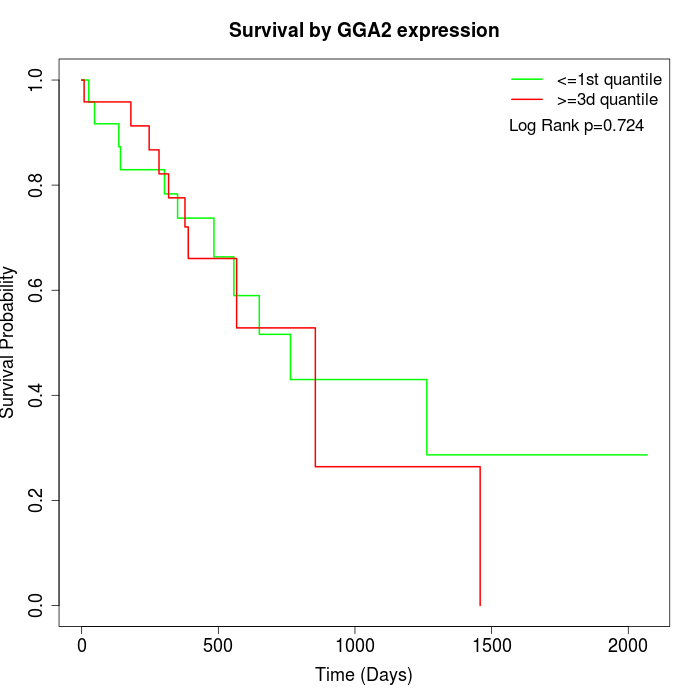
Survival by GGA2 expression:
Note: Click image to view full size file.
Copy number change of GGA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GGA2 | 23062 | 6 | 4 | 20 | |
| GSE20123 | GGA2 | 23062 | 6 | 3 | 21 | |
| GSE43470 | GGA2 | 23062 | 3 | 3 | 37 | |
| GSE46452 | GGA2 | 23062 | 38 | 1 | 20 | |
| GSE47630 | GGA2 | 23062 | 11 | 6 | 23 | |
| GSE54993 | GGA2 | 23062 | 3 | 5 | 62 | |
| GSE54994 | GGA2 | 23062 | 4 | 10 | 39 | |
| GSE60625 | GGA2 | 23062 | 4 | 0 | 7 | |
| GSE74703 | GGA2 | 23062 | 3 | 2 | 31 | |
| GSE74704 | GGA2 | 23062 | 3 | 1 | 16 | |
| TCGA | GGA2 | 23062 | 19 | 13 | 64 |
Total number of gains: 100; Total number of losses: 48; Total Number of normals: 340.
Somatic mutations of GGA2:
Generating mutation plots.
Highly correlated genes for GGA2:
Showing top 20/448 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GGA2 | TTC30B | 0.753158 | 3 | 0 | 3 |
| GGA2 | C9orf16 | 0.739794 | 3 | 0 | 3 |
| GGA2 | ZNF691 | 0.712642 | 3 | 0 | 3 |
| GGA2 | ZNF830 | 0.711693 | 4 | 0 | 4 |
| GGA2 | RBM4B | 0.7097 | 3 | 0 | 3 |
| GGA2 | SCAF1 | 0.706896 | 3 | 0 | 3 |
| GGA2 | RNF166 | 0.700717 | 3 | 0 | 3 |
| GGA2 | RICTOR | 0.699956 | 3 | 0 | 3 |
| GGA2 | RNF114 | 0.697455 | 3 | 0 | 3 |
| GGA2 | GHDC | 0.697218 | 3 | 0 | 3 |
| GGA2 | EEFSEC | 0.696085 | 4 | 0 | 4 |
| GGA2 | CRKL | 0.694257 | 3 | 0 | 3 |
| GGA2 | SEMA4B | 0.688179 | 3 | 0 | 3 |
| GGA2 | IAH1 | 0.687581 | 3 | 0 | 3 |
| GGA2 | EHMT2 | 0.683666 | 4 | 0 | 3 |
| GGA2 | HNRNPAB | 0.682375 | 3 | 0 | 3 |
| GGA2 | PLEKHH1 | 0.681437 | 3 | 0 | 3 |
| GGA2 | INO80E | 0.679386 | 3 | 0 | 3 |
| GGA2 | CDH1 | 0.678271 | 3 | 0 | 3 |
| GGA2 | MTHFSD | 0.677163 | 4 | 0 | 3 |
For details and further investigation, click here