| Full name: ghrelin and obestatin prepropeptide | Alias Symbol: MTLRP|ghrelin|obestatin | ||
| Type: protein-coding gene | Cytoband: 3p25.3 | ||
| Entrez ID: 51738 | HGNC ID: HGNC:18129 | Ensembl Gene: ENSG00000157017 | OMIM ID: 605353 |
| Drug and gene relationship at DGIdb | |||
GHRL involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway |
Expression of GHRL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GHRL | 51738 | 237647_at | 0.0764 | 0.7175 | |
| GSE26886 | GHRL | 51738 | 223862_at | -0.0398 | 0.8099 | |
| GSE45670 | GHRL | 51738 | 237647_at | -0.0536 | 0.6241 | |
| GSE53622 | GHRL | 51738 | 6230 | -1.0385 | 0.0010 | |
| GSE53624 | GHRL | 51738 | 6230 | -1.4200 | 0.0000 | |
| GSE63941 | GHRL | 51738 | 223862_at | -0.0133 | 0.9387 | |
| GSE77861 | GHRL | 51738 | 223862_at | -0.0207 | 0.8332 | |
| GSE97050 | GHRL | 51738 | A_23_P40956 | -0.1343 | 0.8023 | |
| SRP133303 | GHRL | 51738 | RNAseq | -0.9206 | 0.0021 | |
| SRP219564 | GHRL | 51738 | RNAseq | -0.0891 | 0.9033 | |
| TCGA | GHRL | 51738 | RNAseq | -4.6405 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
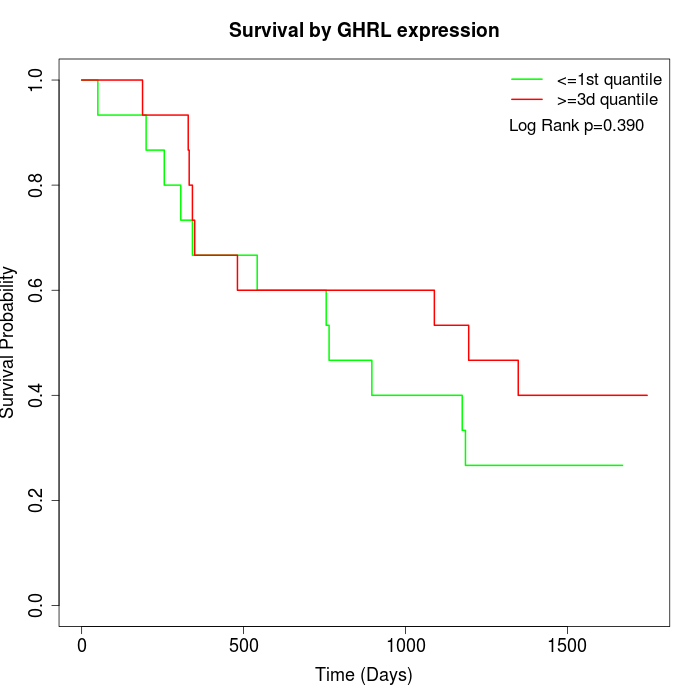
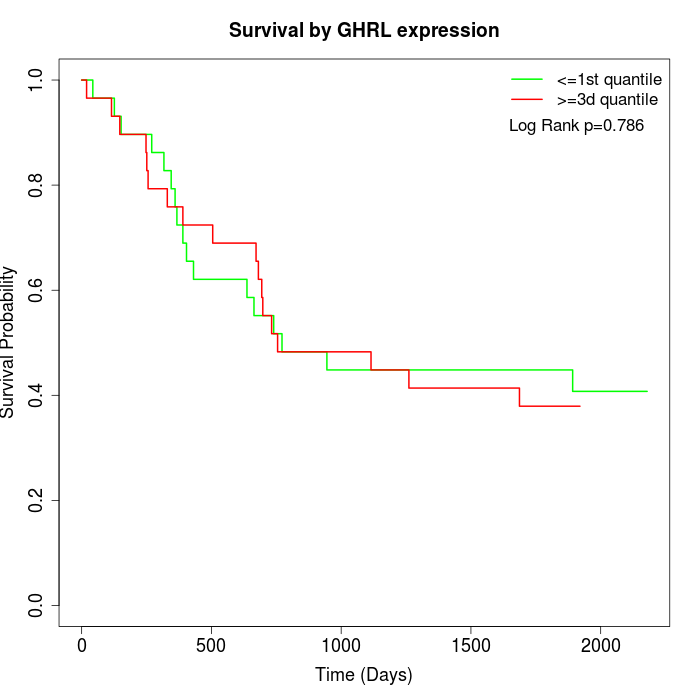
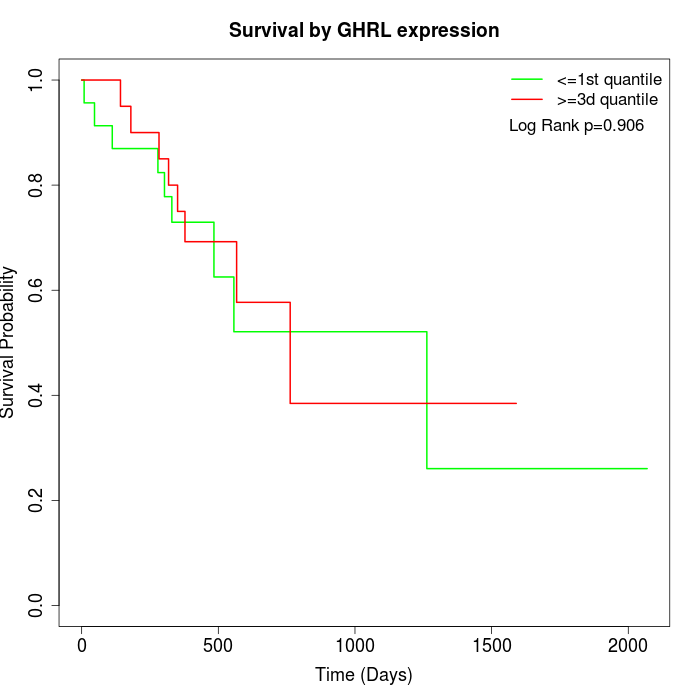
Survival by GHRL expression:
Note: Click image to view full size file.
Copy number change of GHRL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GHRL | 51738 | 1 | 18 | 11 | |
| GSE20123 | GHRL | 51738 | 1 | 18 | 11 | |
| GSE43470 | GHRL | 51738 | 1 | 20 | 22 | |
| GSE46452 | GHRL | 51738 | 2 | 17 | 40 | |
| GSE47630 | GHRL | 51738 | 2 | 23 | 15 | |
| GSE54993 | GHRL | 51738 | 6 | 1 | 63 | |
| GSE54994 | GHRL | 51738 | 3 | 31 | 19 | |
| GSE60625 | GHRL | 51738 | 5 | 0 | 6 | |
| GSE74703 | GHRL | 51738 | 1 | 16 | 19 | |
| GSE74704 | GHRL | 51738 | 1 | 10 | 9 | |
| TCGA | GHRL | 51738 | 4 | 66 | 26 |
Total number of gains: 27; Total number of losses: 220; Total Number of normals: 241.
Somatic mutations of GHRL:
Generating mutation plots.
Highly correlated genes for GHRL:
Showing top 20/25 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GHRL | GKN2 | 0.661673 | 3 | 0 | 3 |
| GHRL | CXCR1 | 0.646507 | 3 | 0 | 3 |
| GHRL | TMPRSS3 | 0.628907 | 4 | 0 | 4 |
| GHRL | FGFRL1 | 0.61205 | 3 | 0 | 3 |
| GHRL | KCNE2 | 0.606887 | 4 | 0 | 3 |
| GHRL | CAPN9 | 0.606026 | 3 | 0 | 3 |
| GHRL | FMO5 | 0.604134 | 4 | 0 | 3 |
| GHRL | DCAF12L1 | 0.602882 | 3 | 0 | 3 |
| GHRL | UGT2B15 | 0.59248 | 3 | 0 | 3 |
| GHRL | PDGFD | 0.58321 | 3 | 0 | 3 |
| GHRL | GABRR2 | 0.578402 | 4 | 0 | 3 |
| GHRL | G0S2 | 0.578349 | 4 | 0 | 4 |
| GHRL | MBOAT4 | 0.578023 | 3 | 0 | 3 |
| GHRL | PROK2 | 0.574748 | 4 | 0 | 3 |
| GHRL | TOX3 | 0.570003 | 4 | 0 | 3 |
| GHRL | TMEM71 | 0.569718 | 4 | 0 | 3 |
| GHRL | GJB1 | 0.566902 | 4 | 0 | 3 |
| GHRL | SLC29A1 | 0.551827 | 3 | 0 | 3 |
| GHRL | SERPINA5 | 0.55136 | 5 | 0 | 3 |
| GHRL | C20orf202 | 0.537214 | 4 | 0 | 3 |
For details and further investigation, click here