| Full name: potassium voltage-gated channel subfamily E regulatory subunit 2 | Alias Symbol: MiRP1|LQT6 | ||
| Type: protein-coding gene | Cytoband: 21q22.11 | ||
| Entrez ID: 9992 | HGNC ID: HGNC:6242 | Ensembl Gene: ENSG00000159197 | OMIM ID: 603796 |
| Drug and gene relationship at DGIdb | |||
KCNE2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04971 | Gastric acid secretion |
Expression of KCNE2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNE2 | 9992 | 221095_s_at | -0.0351 | 0.9218 | |
| GSE20347 | KCNE2 | 9992 | 221095_s_at | 0.0342 | 0.6410 | |
| GSE23400 | KCNE2 | 9992 | 221095_s_at | -0.1009 | 0.0030 | |
| GSE26886 | KCNE2 | 9992 | 221095_s_at | -0.0586 | 0.6199 | |
| GSE29001 | KCNE2 | 9992 | 221095_s_at | -0.1245 | 0.2582 | |
| GSE38129 | KCNE2 | 9992 | 221095_s_at | -0.4602 | 0.1392 | |
| GSE45670 | KCNE2 | 9992 | 221095_s_at | -0.0446 | 0.7245 | |
| GSE53622 | KCNE2 | 9992 | 111938 | -0.2807 | 0.2595 | |
| GSE53624 | KCNE2 | 9992 | 111938 | -0.2045 | 0.3469 | |
| GSE63941 | KCNE2 | 9992 | 221095_s_at | 0.5043 | 0.0129 | |
| GSE77861 | KCNE2 | 9992 | 221095_s_at | -0.0905 | 0.3749 | |
| TCGA | KCNE2 | 9992 | RNAseq | -4.3782 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
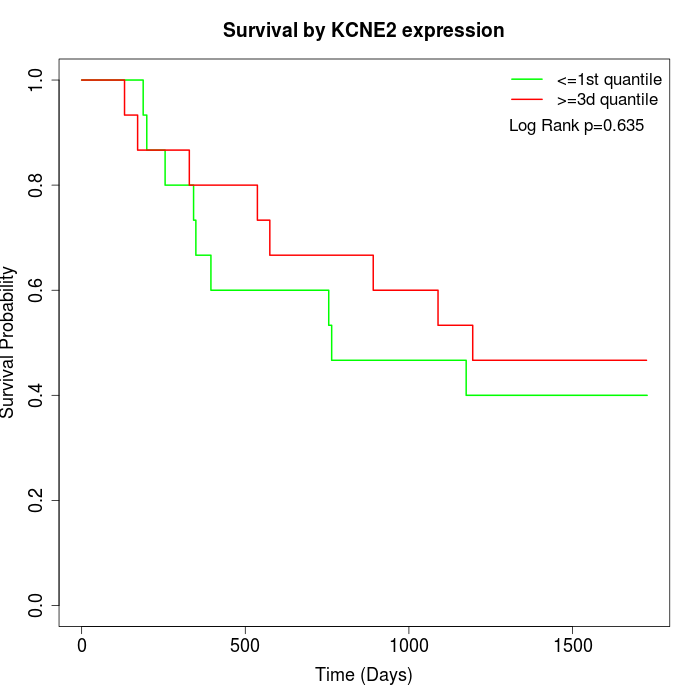
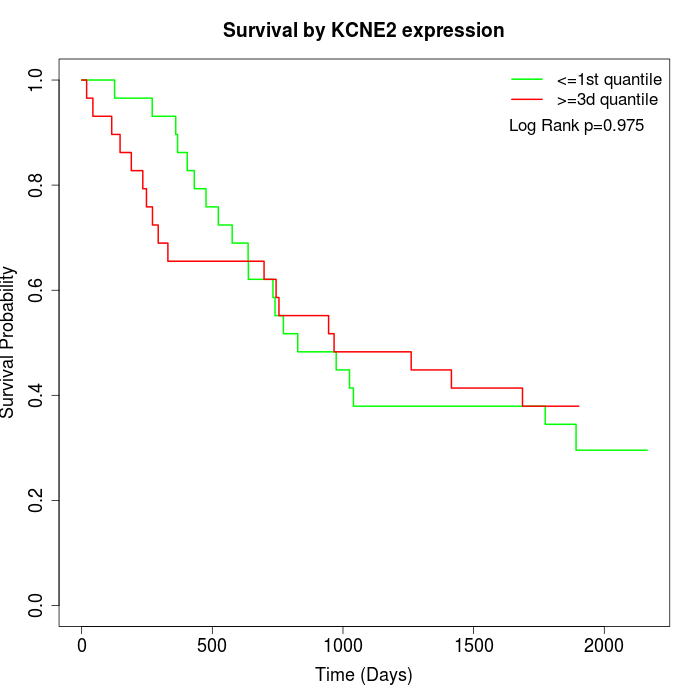
Survival by KCNE2 expression:
Note: Click image to view full size file.
Copy number change of KCNE2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNE2 | 9992 | 2 | 7 | 21 | |
| GSE20123 | KCNE2 | 9992 | 2 | 8 | 20 | |
| GSE43470 | KCNE2 | 9992 | 1 | 11 | 31 | |
| GSE46452 | KCNE2 | 9992 | 1 | 21 | 37 | |
| GSE47630 | KCNE2 | 9992 | 6 | 17 | 17 | |
| GSE54993 | KCNE2 | 9992 | 9 | 1 | 60 | |
| GSE54994 | KCNE2 | 9992 | 1 | 9 | 43 | |
| GSE60625 | KCNE2 | 9992 | 0 | 0 | 11 | |
| GSE74703 | KCNE2 | 9992 | 1 | 8 | 27 | |
| GSE74704 | KCNE2 | 9992 | 1 | 4 | 15 | |
| TCGA | KCNE2 | 9992 | 8 | 37 | 51 |
Total number of gains: 32; Total number of losses: 123; Total Number of normals: 333.
Somatic mutations of KCNE2:
Generating mutation plots.
Highly correlated genes for KCNE2:
Showing top 20/431 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNE2 | GATA6-AS1 | 0.744247 | 3 | 0 | 3 |
| KCNE2 | CHIA | 0.739772 | 5 | 0 | 5 |
| KCNE2 | PDIA2 | 0.734571 | 4 | 0 | 4 |
| KCNE2 | NR0B2 | 0.730848 | 5 | 0 | 5 |
| KCNE2 | CLDN18 | 0.725544 | 5 | 0 | 5 |
| KCNE2 | PGC | 0.707539 | 5 | 0 | 4 |
| KCNE2 | NMNAT2 | 0.684852 | 4 | 0 | 4 |
| KCNE2 | CGA | 0.681263 | 4 | 0 | 3 |
| KCNE2 | OBSL1 | 0.679053 | 3 | 0 | 3 |
| KCNE2 | EPS8L3 | 0.677304 | 6 | 0 | 5 |
| KCNE2 | FRAS1 | 0.674283 | 3 | 0 | 3 |
| KCNE2 | LGALS4 | 0.670474 | 5 | 0 | 4 |
| KCNE2 | ADAM21 | 0.668623 | 3 | 0 | 3 |
| KCNE2 | SERPINA4 | 0.661623 | 7 | 0 | 6 |
| KCNE2 | ATP4A | 0.657566 | 8 | 0 | 5 |
| KCNE2 | SMIM24 | 0.65646 | 3 | 0 | 3 |
| KCNE2 | ASCL1 | 0.654818 | 3 | 0 | 3 |
| KCNE2 | GPR22 | 0.652241 | 3 | 0 | 3 |
| KCNE2 | CPA2 | 0.651971 | 7 | 0 | 5 |
| KCNE2 | DLX6 | 0.651431 | 3 | 0 | 3 |
For details and further investigation, click here