| Full name: C-X-C motif chemokine receptor 1 | Alias Symbol: CKR-1|CDw128a|CD181 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 3577 | HGNC ID: HGNC:6026 | Ensembl Gene: ENSG00000163464 | OMIM ID: 146929 |
| Drug and gene relationship at DGIdb | |||
CXCR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04072 | Phospholipase D signaling pathway | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of CXCR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCR1 | 3577 | 207094_at | 0.1779 | 0.7765 | |
| GSE20347 | CXCR1 | 3577 | 207094_at | -0.0618 | 0.3795 | |
| GSE23400 | CXCR1 | 3577 | 207094_at | -0.0898 | 0.0130 | |
| GSE26886 | CXCR1 | 3577 | 207094_at | -0.1973 | 0.1528 | |
| GSE29001 | CXCR1 | 3577 | 207094_at | -0.0857 | 0.5939 | |
| GSE38129 | CXCR1 | 3577 | 207094_at | -0.1464 | 0.0193 | |
| GSE45670 | CXCR1 | 3577 | 207094_at | 0.2326 | 0.0280 | |
| GSE53622 | CXCR1 | 3577 | 97782 | 0.3908 | 0.0905 | |
| GSE53624 | CXCR1 | 3577 | 97782 | 0.4476 | 0.0287 | |
| GSE63941 | CXCR1 | 3577 | 207094_at | 0.0132 | 0.9513 | |
| GSE77861 | CXCR1 | 3577 | 207094_at | 0.0317 | 0.7397 | |
| GSE97050 | CXCR1 | 3577 | A_23_P67932 | -0.1397 | 0.8180 | |
| SRP133303 | CXCR1 | 3577 | RNAseq | 1.8544 | 0.0000 | |
| TCGA | CXCR1 | 3577 | RNAseq | -0.0990 | 0.8486 |
Upregulated datasets: 1; Downregulated datasets: 0.
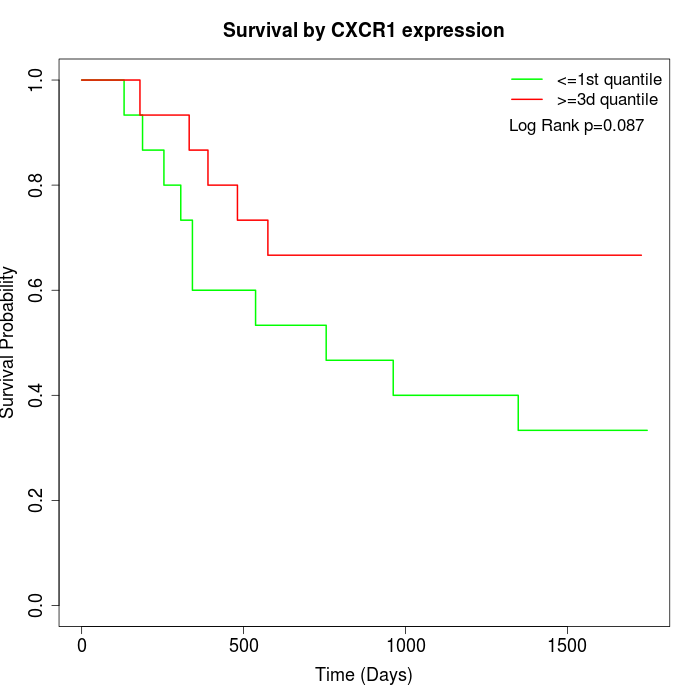
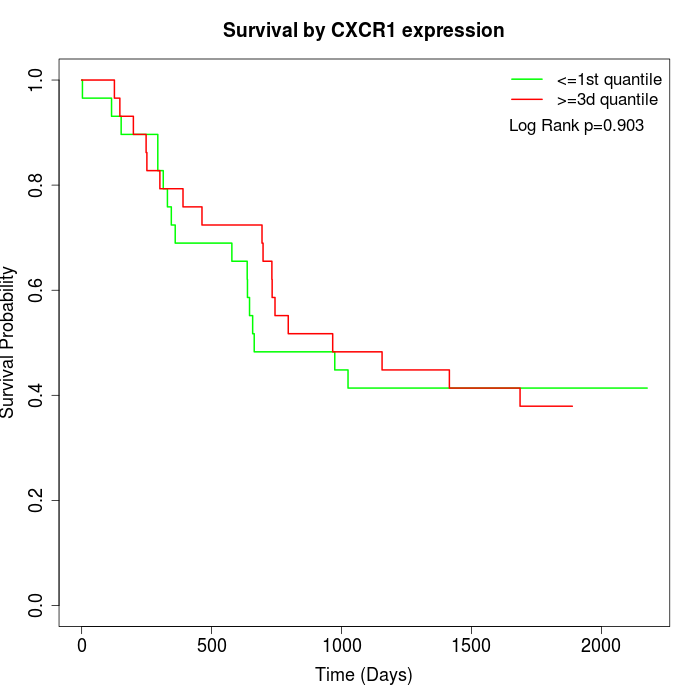
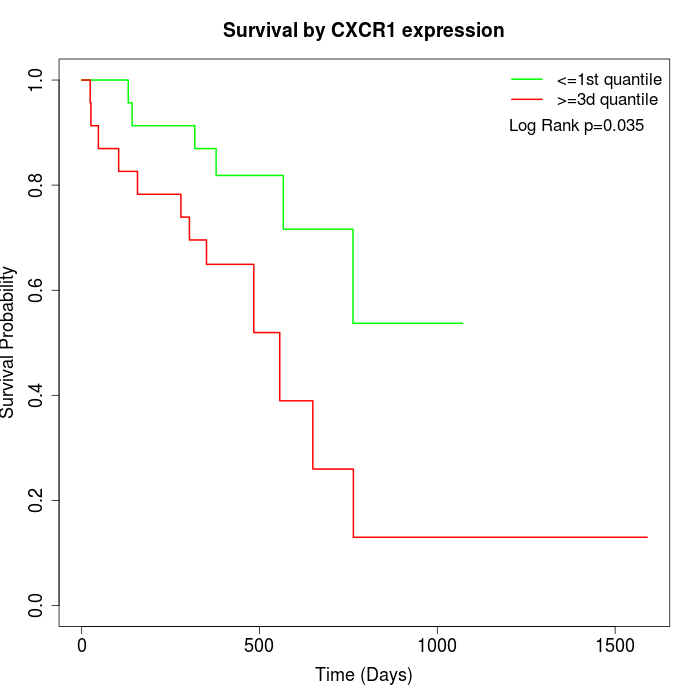
Survival by CXCR1 expression:
Note: Click image to view full size file.
Copy number change of CXCR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCR1 | 3577 | 1 | 13 | 16 | |
| GSE20123 | CXCR1 | 3577 | 1 | 13 | 16 | |
| GSE43470 | CXCR1 | 3577 | 1 | 7 | 35 | |
| GSE46452 | CXCR1 | 3577 | 0 | 5 | 54 | |
| GSE47630 | CXCR1 | 3577 | 4 | 5 | 31 | |
| GSE54993 | CXCR1 | 3577 | 1 | 2 | 67 | |
| GSE54994 | CXCR1 | 3577 | 8 | 10 | 35 | |
| GSE60625 | CXCR1 | 3577 | 0 | 3 | 8 | |
| GSE74703 | CXCR1 | 3577 | 1 | 5 | 30 | |
| GSE74704 | CXCR1 | 3577 | 1 | 7 | 12 | |
| TCGA | CXCR1 | 3577 | 11 | 27 | 58 |
Total number of gains: 29; Total number of losses: 97; Total Number of normals: 362.
Somatic mutations of CXCR1:
Generating mutation plots.
Highly correlated genes for CXCR1:
Showing top 20/782 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCR1 | ANHX | 0.770142 | 3 | 0 | 3 |
| CXCR1 | CMTM2 | 0.745265 | 5 | 0 | 4 |
| CXCR1 | SYT13 | 0.737473 | 3 | 0 | 3 |
| CXCR1 | PHLDB1 | 0.734356 | 5 | 0 | 5 |
| CXCR1 | GCNT2 | 0.715993 | 3 | 0 | 3 |
| CXCR1 | SOX5 | 0.708768 | 3 | 0 | 3 |
| CXCR1 | NOS2 | 0.70303 | 4 | 0 | 3 |
| CXCR1 | CDH15 | 0.697772 | 5 | 0 | 5 |
| CXCR1 | PACS2 | 0.694227 | 3 | 0 | 3 |
| CXCR1 | PROK2 | 0.686595 | 6 | 0 | 5 |
| CXCR1 | DNALI1 | 0.685218 | 3 | 0 | 3 |
| CXCR1 | CISH | 0.683935 | 6 | 0 | 6 |
| CXCR1 | C20orf202 | 0.681682 | 4 | 0 | 3 |
| CXCR1 | STAC | 0.679988 | 4 | 0 | 3 |
| CXCR1 | CA8 | 0.679871 | 3 | 0 | 3 |
| CXCR1 | KIR2DS2 | 0.678835 | 5 | 0 | 5 |
| CXCR1 | NFAM1 | 0.677482 | 3 | 0 | 3 |
| CXCR1 | SLC22A18AS | 0.675463 | 4 | 0 | 4 |
| CXCR1 | NEUROG3 | 0.674067 | 5 | 0 | 5 |
| CXCR1 | GPR22 | 0.671783 | 3 | 0 | 3 |
For details and further investigation, click here