| Full name: prokineticin 2 | Alias Symbol: PK2|BV8|MIT1|KAL4 | ||
| Type: protein-coding gene | Cytoband: 3p13 | ||
| Entrez ID: 60675 | HGNC ID: HGNC:18455 | Ensembl Gene: ENSG00000163421 | OMIM ID: 607002 |
| Drug and gene relationship at DGIdb | |||
Expression of PROK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PROK2 | 60675 | 232629_at | 1.0773 | 0.3040 | |
| GSE26886 | PROK2 | 60675 | 232629_at | -0.0194 | 0.8762 | |
| GSE45670 | PROK2 | 60675 | 232629_at | 0.4024 | 0.4379 | |
| GSE53622 | PROK2 | 60675 | 42883 | 0.6873 | 0.0609 | |
| GSE53624 | PROK2 | 60675 | 42883 | 0.7321 | 0.0196 | |
| GSE63941 | PROK2 | 60675 | 232629_at | 0.1203 | 0.3044 | |
| GSE77861 | PROK2 | 60675 | 232629_at | 0.0587 | 0.5928 | |
| GSE97050 | PROK2 | 60675 | A_24_P97342 | -0.1676 | 0.7363 | |
| TCGA | PROK2 | 60675 | RNAseq | -0.1091 | 0.8661 |
Upregulated datasets: 0; Downregulated datasets: 0.
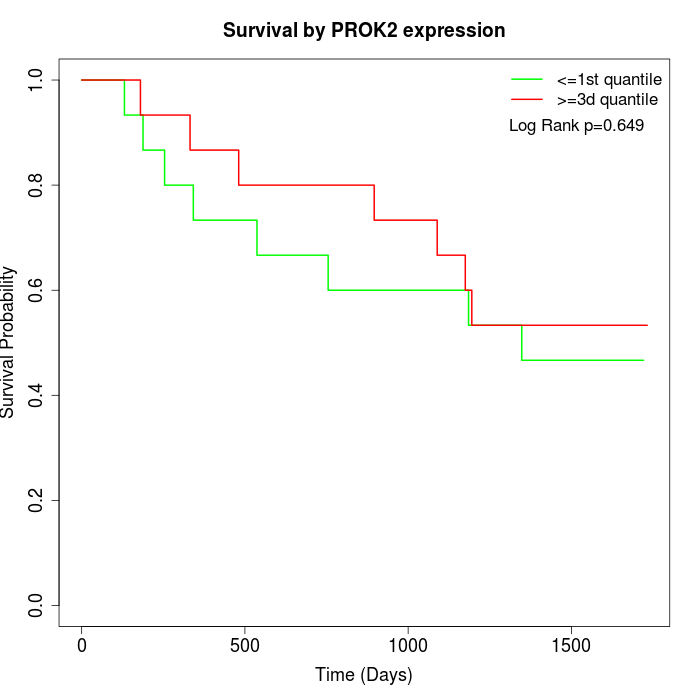
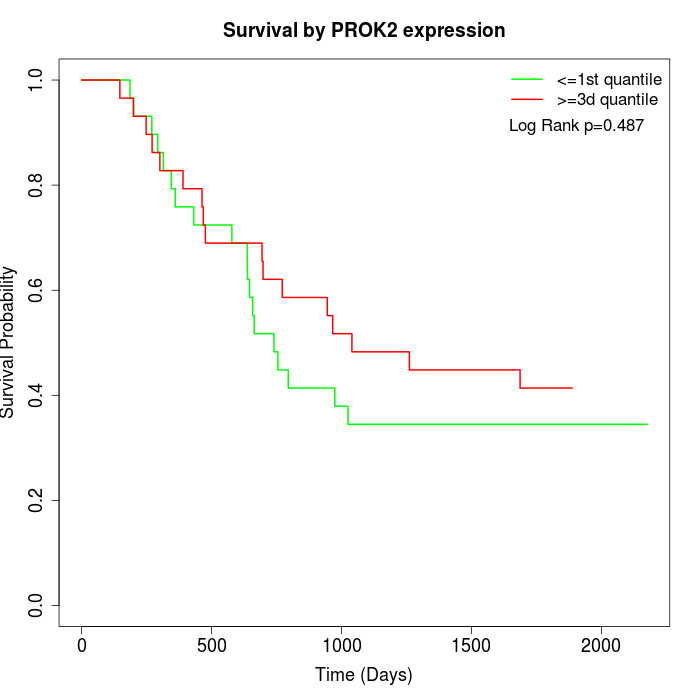
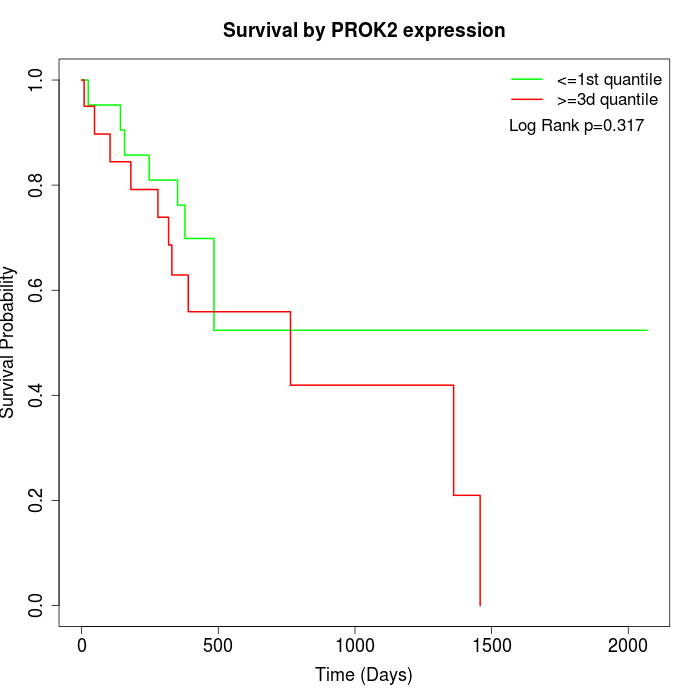
Survival by PROK2 expression:
Note: Click image to view full size file.
Copy number change of PROK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PROK2 | 60675 | 1 | 20 | 9 | |
| GSE20123 | PROK2 | 60675 | 1 | 20 | 9 | |
| GSE43470 | PROK2 | 60675 | 0 | 21 | 22 | |
| GSE46452 | PROK2 | 60675 | 2 | 17 | 40 | |
| GSE47630 | PROK2 | 60675 | 1 | 23 | 16 | |
| GSE54993 | PROK2 | 60675 | 5 | 3 | 62 | |
| GSE54994 | PROK2 | 60675 | 1 | 33 | 19 | |
| GSE60625 | PROK2 | 60675 | 3 | 1 | 7 | |
| GSE74703 | PROK2 | 60675 | 0 | 17 | 19 | |
| GSE74704 | PROK2 | 60675 | 1 | 13 | 6 | |
| TCGA | PROK2 | 60675 | 1 | 79 | 16 |
Total number of gains: 16; Total number of losses: 247; Total Number of normals: 225.
Somatic mutations of PROK2:
Generating mutation plots.
Highly correlated genes for PROK2:
Showing top 20/154 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PROK2 | TNFRSF10C | 0.830549 | 3 | 0 | 3 |
| PROK2 | CMTM2 | 0.825001 | 5 | 0 | 5 |
| PROK2 | FOXC2 | 0.762919 | 3 | 0 | 3 |
| PROK2 | FCGR3B | 0.758097 | 4 | 0 | 4 |
| PROK2 | CERK | 0.753452 | 3 | 0 | 3 |
| PROK2 | BCL2A1 | 0.746522 | 6 | 0 | 6 |
| PROK2 | FAM167B | 0.739396 | 3 | 0 | 3 |
| PROK2 | AVIL | 0.727057 | 3 | 0 | 3 |
| PROK2 | CSF3R | 0.719693 | 5 | 0 | 4 |
| PROK2 | PADI4 | 0.71728 | 3 | 0 | 3 |
| PROK2 | IL3RA | 0.717213 | 3 | 0 | 3 |
| PROK2 | IL13RA2 | 0.716969 | 5 | 0 | 5 |
| PROK2 | CXCL5 | 0.705388 | 5 | 0 | 5 |
| PROK2 | AQP9 | 0.688884 | 8 | 0 | 6 |
| PROK2 | CXCR1 | 0.686595 | 6 | 0 | 5 |
| PROK2 | PJA1 | 0.686531 | 4 | 0 | 4 |
| PROK2 | NFE2 | 0.681026 | 5 | 0 | 5 |
| PROK2 | RHOBTB2 | 0.66907 | 3 | 0 | 3 |
| PROK2 | MGAT5 | 0.668882 | 4 | 0 | 3 |
| PROK2 | ATP6V1B2 | 0.666718 | 3 | 0 | 3 |
For details and further investigation, click here