| Full name: GNAS complex locus | Alias Symbol: NESP55|NESP|GNASXL|GPSA|SCG6|SgVI | ||
| Type: protein-coding gene | Cytoband: 20q13.32 | ||
| Entrez ID: 2778 | HGNC ID: HGNC:4392 | Ensembl Gene: ENSG00000087460 | OMIM ID: 139320 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
GNAS involved pathways:
Expression of GNAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNAS | 2778 | 200780_x_at | 0.2660 | 0.3470 | |
| GSE20347 | GNAS | 2778 | 200780_x_at | -0.1194 | 0.3405 | |
| GSE23400 | GNAS | 2778 | 214548_x_at | 0.0646 | 0.2910 | |
| GSE26886 | GNAS | 2778 | 200780_x_at | 0.2869 | 0.1283 | |
| GSE29001 | GNAS | 2778 | 200981_x_at | -0.0679 | 0.8706 | |
| GSE38129 | GNAS | 2778 | 200780_x_at | -0.0541 | 0.6420 | |
| GSE45670 | GNAS | 2778 | 200780_x_at | -0.2401 | 0.0092 | |
| GSE53622 | GNAS | 2778 | 47139 | 0.2321 | 0.0008 | |
| GSE53624 | GNAS | 2778 | 47139 | 0.1356 | 0.0059 | |
| GSE63941 | GNAS | 2778 | 200780_x_at | 0.5530 | 0.1165 | |
| GSE77861 | GNAS | 2778 | 200780_x_at | -0.2777 | 0.2088 | |
| GSE97050 | GNAS | 2778 | A_24_P418809 | -0.0590 | 0.8390 | |
| SRP007169 | GNAS | 2778 | RNAseq | -1.1413 | 0.0065 | |
| SRP008496 | GNAS | 2778 | RNAseq | -0.9067 | 0.0001 | |
| SRP064894 | GNAS | 2778 | RNAseq | -0.0779 | 0.7032 | |
| SRP133303 | GNAS | 2778 | RNAseq | -0.1408 | 0.2685 | |
| SRP159526 | GNAS | 2778 | RNAseq | 0.5932 | 0.1894 | |
| SRP193095 | GNAS | 2778 | RNAseq | -0.3455 | 0.0013 | |
| SRP219564 | GNAS | 2778 | RNAseq | -0.1980 | 0.6462 | |
| TCGA | GNAS | 2778 | RNAseq | -0.0230 | 0.5056 |
Upregulated datasets: 0; Downregulated datasets: 1.
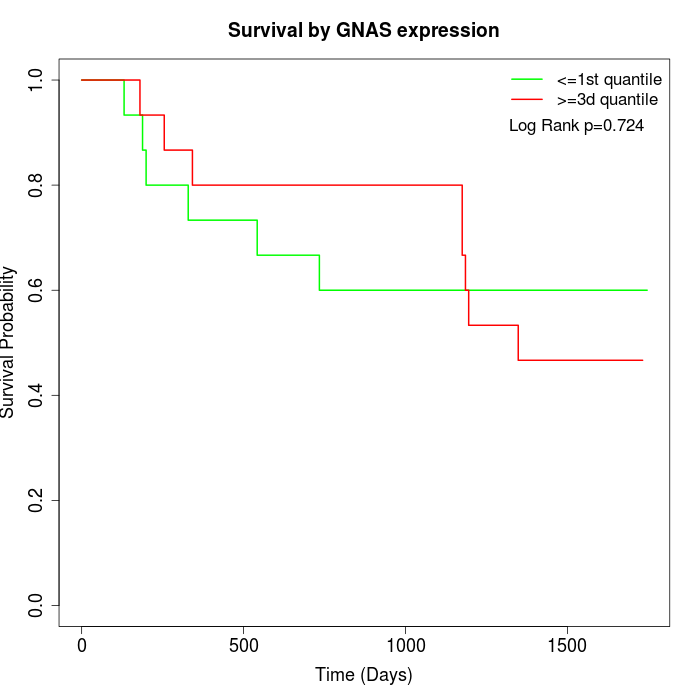
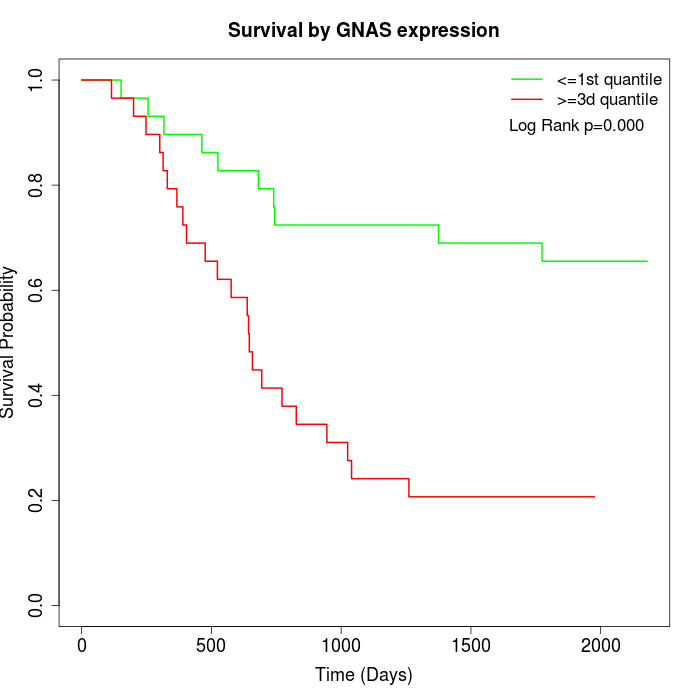
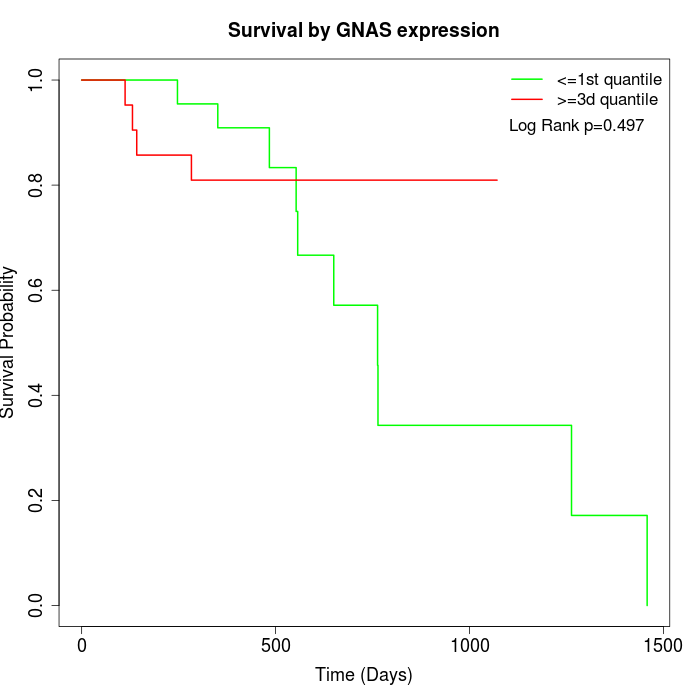
Survival by GNAS expression:
Note: Click image to view full size file.
Copy number change of GNAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNAS | 2778 | 14 | 2 | 14 | |
| GSE20123 | GNAS | 2778 | 14 | 2 | 14 | |
| GSE43470 | GNAS | 2778 | 13 | 0 | 30 | |
| GSE46452 | GNAS | 2778 | 29 | 0 | 30 | |
| GSE47630 | GNAS | 2778 | 24 | 1 | 15 | |
| GSE54993 | GNAS | 2778 | 0 | 17 | 53 | |
| GSE54994 | GNAS | 2778 | 29 | 0 | 24 | |
| GSE60625 | GNAS | 2778 | 0 | 0 | 11 | |
| GSE74703 | GNAS | 2778 | 11 | 0 | 25 | |
| GSE74704 | GNAS | 2778 | 11 | 0 | 9 | |
| TCGA | GNAS | 2778 | 43 | 4 | 49 |
Total number of gains: 188; Total number of losses: 26; Total Number of normals: 274.
Somatic mutations of GNAS:
Generating mutation plots.
Highly correlated genes for GNAS:
Showing top 20/171 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNAS | HDAC6 | 0.771656 | 3 | 0 | 3 |
| GNAS | ZNF213 | 0.764352 | 3 | 0 | 3 |
| GNAS | ZGPAT | 0.730959 | 5 | 0 | 5 |
| GNAS | NGF | 0.71844 | 3 | 0 | 3 |
| GNAS | POLR2L | 0.715665 | 3 | 0 | 3 |
| GNAS | XRRA1 | 0.709579 | 3 | 0 | 3 |
| GNAS | PYGO1 | 0.708218 | 3 | 0 | 3 |
| GNAS | C1orf35 | 0.706573 | 3 | 0 | 3 |
| GNAS | PIP4K2B | 0.706151 | 3 | 0 | 3 |
| GNAS | GALM | 0.694426 | 3 | 0 | 3 |
| GNAS | GUSB | 0.692154 | 3 | 0 | 3 |
| GNAS | BCL9L | 0.688437 | 3 | 0 | 3 |
| GNAS | ACIN1 | 0.677257 | 3 | 0 | 3 |
| GNAS | RRM2B | 0.664406 | 3 | 0 | 3 |
| GNAS | MRPL55 | 0.662041 | 3 | 0 | 3 |
| GNAS | MED25 | 0.658765 | 3 | 0 | 3 |
| GNAS | MAF1 | 0.650356 | 4 | 0 | 3 |
| GNAS | ROGDI | 0.647835 | 3 | 0 | 3 |
| GNAS | MED29 | 0.645136 | 3 | 0 | 3 |
| GNAS | ZNF503 | 0.641402 | 4 | 0 | 4 |
For details and further investigation, click here