| Full name: nerve growth factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 4803 | HGNC ID: HGNC:7808 | Ensembl Gene: ENSG00000134259 | OMIM ID: 162030 |
| Drug and gene relationship at DGIdb | |||
NGF involved pathways:
Expression of NGF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NGF | 4803 | 206814_at | 0.2201 | 0.4131 | |
| GSE20347 | NGF | 4803 | 206814_at | 0.0985 | 0.2994 | |
| GSE23400 | NGF | 4803 | 206814_at | -0.0485 | 0.2647 | |
| GSE26886 | NGF | 4803 | 206814_at | 0.4699 | 0.0001 | |
| GSE29001 | NGF | 4803 | 206814_at | -0.0484 | 0.8172 | |
| GSE38129 | NGF | 4803 | 206814_at | 0.0445 | 0.7138 | |
| GSE45670 | NGF | 4803 | 206814_at | -0.0660 | 0.5687 | |
| GSE63941 | NGF | 4803 | 206814_at | 0.0724 | 0.8810 | |
| GSE77861 | NGF | 4803 | 206814_at | 0.0460 | 0.8330 | |
| GSE97050 | NGF | 4803 | A_23_P115190 | 0.1412 | 0.6137 | |
| TCGA | NGF | 4803 | RNAseq | 0.8528 | 0.0420 |
Upregulated datasets: 0; Downregulated datasets: 0.
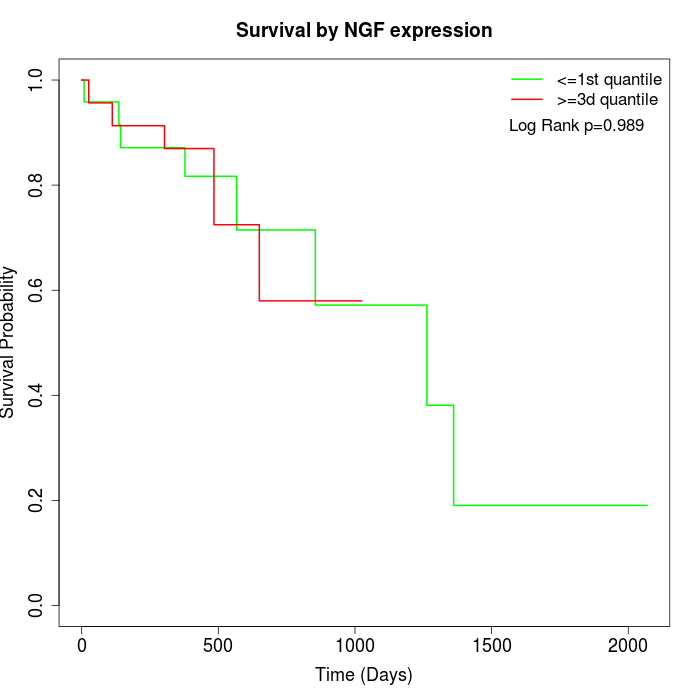
Survival by NGF expression:
Note: Click image to view full size file.
Copy number change of NGF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NGF | 4803 | 0 | 9 | 21 | |
| GSE20123 | NGF | 4803 | 0 | 9 | 21 | |
| GSE43470 | NGF | 4803 | 0 | 8 | 35 | |
| GSE46452 | NGF | 4803 | 2 | 1 | 56 | |
| GSE47630 | NGF | 4803 | 9 | 5 | 26 | |
| GSE54993 | NGF | 4803 | 0 | 1 | 69 | |
| GSE54994 | NGF | 4803 | 8 | 3 | 42 | |
| GSE60625 | NGF | 4803 | 0 | 0 | 11 | |
| GSE74703 | NGF | 4803 | 0 | 7 | 29 | |
| GSE74704 | NGF | 4803 | 0 | 5 | 15 | |
| TCGA | NGF | 4803 | 12 | 29 | 55 |
Total number of gains: 31; Total number of losses: 77; Total Number of normals: 380.
Somatic mutations of NGF:
Generating mutation plots.
Highly correlated genes for NGF:
Showing top 20/538 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NGF | SAFB2 | 0.776196 | 3 | 0 | 3 |
| NGF | TNFRSF12A | 0.763633 | 3 | 0 | 3 |
| NGF | A4GALT | 0.763416 | 3 | 0 | 3 |
| NGF | BRAT1 | 0.762911 | 3 | 0 | 3 |
| NGF | FAM89B | 0.761941 | 3 | 0 | 3 |
| NGF | ISM2 | 0.752662 | 3 | 0 | 3 |
| NGF | POLR2J | 0.750454 | 3 | 0 | 3 |
| NGF | C11orf24 | 0.747717 | 3 | 0 | 3 |
| NGF | TRIM47 | 0.747078 | 3 | 0 | 3 |
| NGF | ABHD6 | 0.746898 | 3 | 0 | 3 |
| NGF | CCS | 0.745632 | 3 | 0 | 3 |
| NGF | BRI3 | 0.744511 | 3 | 0 | 3 |
| NGF | STC2 | 0.744016 | 4 | 0 | 4 |
| NGF | DOCK5 | 0.741174 | 3 | 0 | 3 |
| NGF | MYC | 0.738448 | 3 | 0 | 3 |
| NGF | MARK1 | 0.735248 | 3 | 0 | 3 |
| NGF | SH3RF3 | 0.734288 | 3 | 0 | 3 |
| NGF | NRP2 | 0.733657 | 3 | 0 | 3 |
| NGF | FOXC2 | 0.733155 | 4 | 0 | 3 |
| NGF | FZD4 | 0.733068 | 3 | 0 | 3 |
For details and further investigation, click here