| Full name: golgin A6 family like 9 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 15q25.2 | ||
| Entrez ID: 440295 | HGNC ID: HGNC:37229 | Ensembl Gene: ENSG00000197978 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GOLGA6L9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | GOLGA6L9 | 440295 | 59187 | -0.5045 | 0.0000 | |
| GSE53624 | GOLGA6L9 | 440295 | 59187 | -0.6822 | 0.0000 | |
| GSE97050 | GOLGA6L9 | 440295 | A_33_P3243878 | -0.5949 | 0.1512 | |
| SRP007169 | GOLGA6L9 | 440295 | RNAseq | -0.8084 | 0.1287 | |
| SRP064894 | GOLGA6L9 | 440295 | RNAseq | -1.8482 | 0.0000 | |
| SRP133303 | GOLGA6L9 | 440295 | RNAseq | -0.9857 | 0.0000 | |
| SRP159526 | GOLGA6L9 | 440295 | RNAseq | -1.3676 | 0.0007 | |
| SRP193095 | GOLGA6L9 | 440295 | RNAseq | -1.1073 | 0.0000 | |
| SRP219564 | GOLGA6L9 | 440295 | RNAseq | -1.1845 | 0.1241 | |
| TCGA | GOLGA6L9 | 440295 | RNAseq | -0.1693 | 0.0857 |
Upregulated datasets: 0; Downregulated datasets: 3.
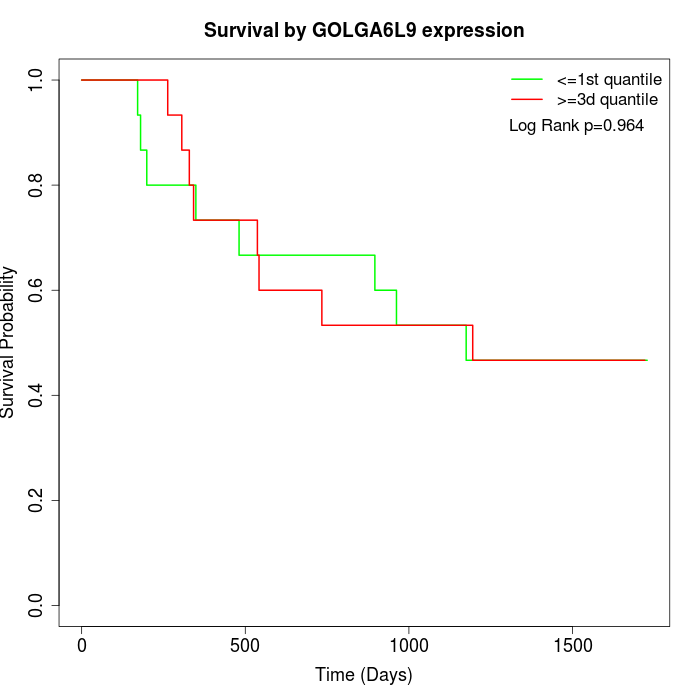
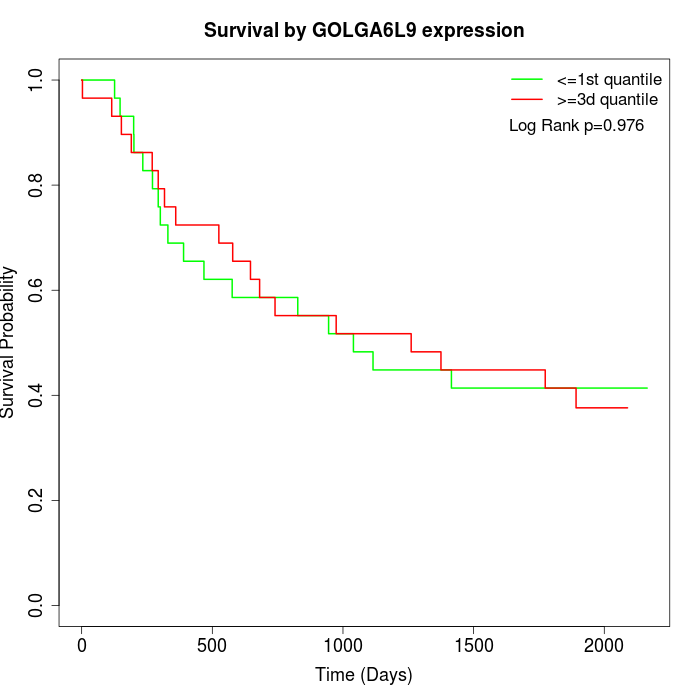
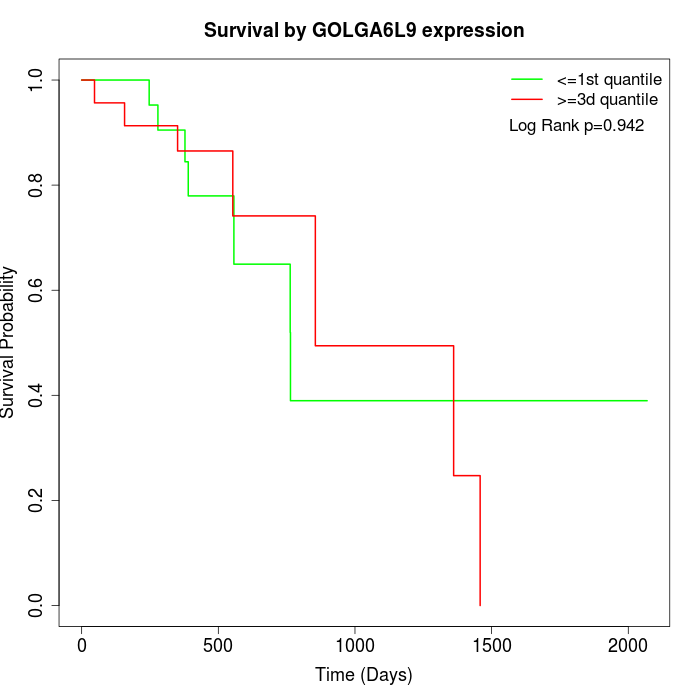
Survival by GOLGA6L9 expression:
Note: Click image to view full size file.
Copy number change of GOLGA6L9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GOLGA6L9 | 440295 | 7 | 1 | 22 | |
| GSE20123 | GOLGA6L9 | 440295 | 7 | 1 | 22 | |
| GSE43470 | GOLGA6L9 | 440295 | 4 | 4 | 35 | |
| GSE46452 | GOLGA6L9 | 440295 | 3 | 7 | 49 | |
| GSE47630 | GOLGA6L9 | 440295 | 8 | 10 | 22 | |
| GSE54993 | GOLGA6L9 | 440295 | 5 | 6 | 59 | |
| GSE54994 | GOLGA6L9 | 440295 | 5 | 7 | 41 | |
| GSE60625 | GOLGA6L9 | 440295 | 4 | 0 | 7 | |
| GSE74703 | GOLGA6L9 | 440295 | 4 | 2 | 30 | |
| GSE74704 | GOLGA6L9 | 440295 | 3 | 1 | 16 | |
| TCGA | GOLGA6L9|ENSG00000196648.6 | NA | 15 | 14 | 67 |
Total number of gains: 65; Total number of losses: 53; Total Number of normals: 370.
Somatic mutations of GOLGA6L9:
Generating mutation plots.
Highly correlated genes for GOLGA6L9:
Showing top 20/38 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GOLGA6L9 | GOLGA6L10 | 0.868576 | 3 | 0 | 3 |
| GOLGA6L9 | MTMR10 | 0.721231 | 3 | 0 | 3 |
| GOLGA6L9 | PLXNB1 | 0.709515 | 3 | 0 | 3 |
| GOLGA6L9 | ECHDC2 | 0.671143 | 3 | 0 | 3 |
| GOLGA6L9 | NBPF11 | 0.666039 | 3 | 0 | 3 |
| GOLGA6L9 | ZDHHC21 | 0.659337 | 3 | 0 | 3 |
| GOLGA6L9 | ARF5 | 0.657076 | 3 | 0 | 3 |
| GOLGA6L9 | ABCA5 | 0.651647 | 3 | 0 | 3 |
| GOLGA6L9 | GOLGA8A | 0.649121 | 3 | 0 | 3 |
| GOLGA6L9 | NDST1 | 0.647625 | 3 | 0 | 3 |
| GOLGA6L9 | CUEDC1 | 0.647539 | 3 | 0 | 3 |
| GOLGA6L9 | TRIP10 | 0.646955 | 3 | 0 | 3 |
| GOLGA6L9 | SECISBP2L | 0.644559 | 3 | 0 | 3 |
| GOLGA6L9 | TMEM127 | 0.640304 | 3 | 0 | 3 |
| GOLGA6L9 | SBF1 | 0.635089 | 3 | 0 | 3 |
| GOLGA6L9 | RNF208 | 0.617028 | 3 | 0 | 3 |
| GOLGA6L9 | NBPF1 | 0.615251 | 3 | 0 | 3 |
| GOLGA6L9 | PLEKHM1 | 0.613252 | 3 | 0 | 3 |
| GOLGA6L9 | NEIL1 | 0.610534 | 3 | 0 | 3 |
| GOLGA6L9 | INO80D | 0.610397 | 3 | 0 | 3 |
For details and further investigation, click here