| Full name: G protein-coupled receptor 101 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xq26.3 | ||
| Entrez ID: 83550 | HGNC ID: HGNC:14963 | Ensembl Gene: ENSG00000165370 | OMIM ID: 300393 |
| Drug and gene relationship at DGIdb | |||
Expression of GPR101:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR101 | 83550 | 1553544_at | -0.0033 | 0.9943 | |
| GSE26886 | GPR101 | 83550 | 1553544_at | -0.2475 | 0.0329 | |
| GSE45670 | GPR101 | 83550 | 1553544_at | -0.0305 | 0.8034 | |
| GSE53622 | GPR101 | 83550 | 26720 | -0.6185 | 0.0000 | |
| GSE53624 | GPR101 | 83550 | 26720 | -1.0459 | 0.0000 | |
| GSE63941 | GPR101 | 83550 | 1553544_at | -0.0825 | 0.6624 | |
| GSE77861 | GPR101 | 83550 | 1553544_at | -0.2428 | 0.0149 |
Upregulated datasets: 0; Downregulated datasets: 1.
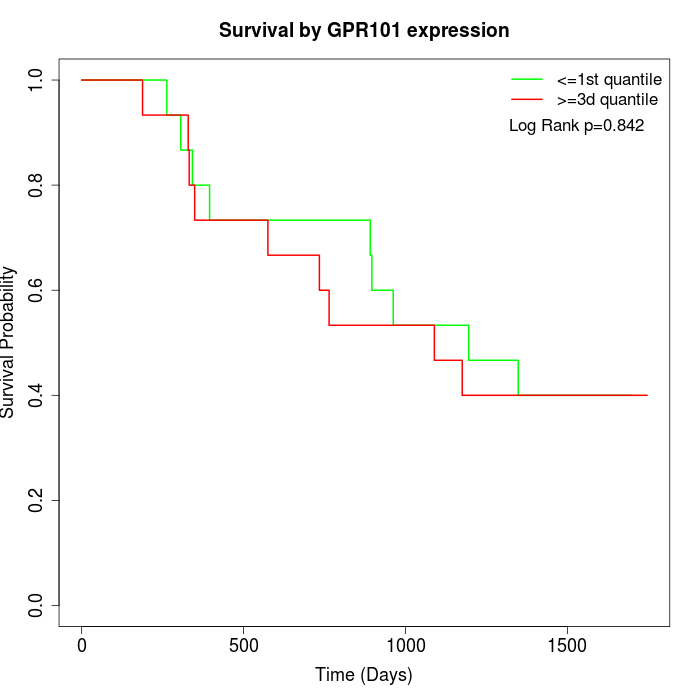
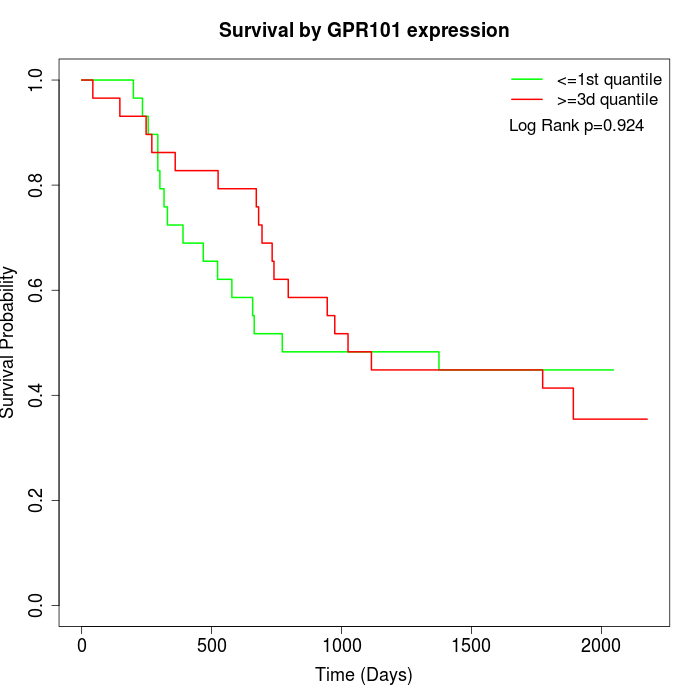
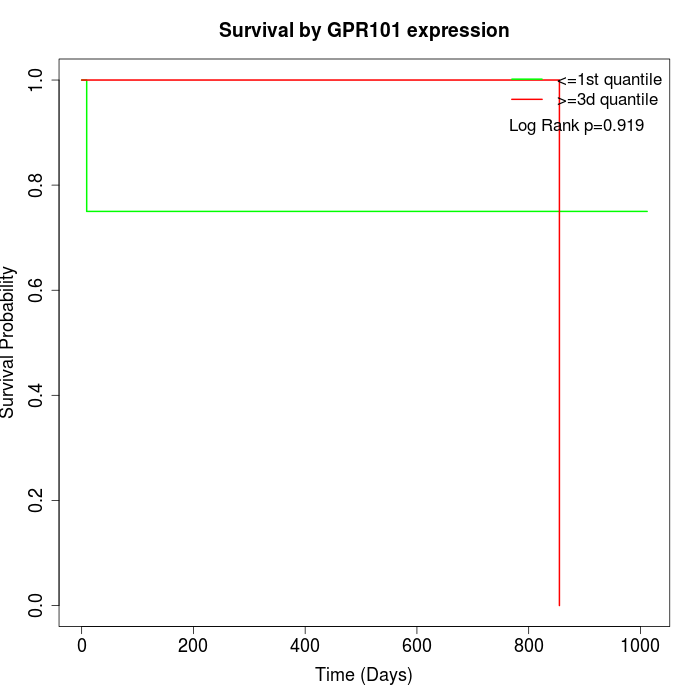
Survival by GPR101 expression:
Note: Click image to view full size file.
Copy number change of GPR101:
No record found for this gene.
Somatic mutations of GPR101:
Generating mutation plots.
Highly correlated genes for GPR101:
Showing top 20/141 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR101 | HDGFL1 | 0.634386 | 4 | 0 | 3 |
| GPR101 | TULP2 | 0.600482 | 5 | 0 | 5 |
| GPR101 | CRCT1 | 0.597601 | 4 | 0 | 3 |
| GPR101 | A1CF | 0.593694 | 3 | 0 | 3 |
| GPR101 | SPATA19 | 0.592454 | 5 | 0 | 4 |
| GPR101 | PKD1L2 | 0.589822 | 3 | 0 | 3 |
| GPR101 | CRABP2 | 0.589342 | 4 | 0 | 4 |
| GPR101 | ENDOU | 0.587546 | 4 | 0 | 4 |
| GPR101 | SBSN | 0.586805 | 4 | 0 | 3 |
| GPR101 | EPS8L2 | 0.581751 | 4 | 0 | 3 |
| GPR101 | FAM209B | 0.581409 | 3 | 0 | 3 |
| GPR101 | NMU | 0.581299 | 3 | 0 | 3 |
| GPR101 | IL17RC | 0.580212 | 5 | 0 | 4 |
| GPR101 | PIM1 | 0.577678 | 5 | 0 | 3 |
| GPR101 | HTR3B | 0.573905 | 5 | 0 | 4 |
| GPR101 | KRT83 | 0.573639 | 3 | 0 | 3 |
| GPR101 | RBM47 | 0.573134 | 4 | 0 | 3 |
| GPR101 | AIF1L | 0.573101 | 4 | 0 | 3 |
| GPR101 | RIIAD1 | 0.571902 | 3 | 0 | 3 |
| GPR101 | GRTP1-AS1 | 0.571545 | 4 | 0 | 3 |
For details and further investigation, click here