| Full name: potassium voltage-gated channel subfamily D member 3 | Alias Symbol: Kv4.3|KSHIVB | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 3752 | HGNC ID: HGNC:6239 | Ensembl Gene: ENSG00000171385 | OMIM ID: 605411 |
| Drug and gene relationship at DGIdb | |||
Expression of KCND3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCND3 | 3752 | 213832_at | -1.0663 | 0.1694 | |
| GSE20347 | KCND3 | 3752 | 211301_at | -0.0397 | 0.5909 | |
| GSE23400 | KCND3 | 3752 | 211301_at | -0.0653 | 0.0723 | |
| GSE26886 | KCND3 | 3752 | 211301_at | 0.3767 | 0.0200 | |
| GSE29001 | KCND3 | 3752 | 211301_at | -0.0589 | 0.7479 | |
| GSE38129 | KCND3 | 3752 | 213832_at | -1.1985 | 0.0003 | |
| GSE45670 | KCND3 | 3752 | 213832_at | -0.8245 | 0.0356 | |
| GSE53622 | KCND3 | 3752 | 44020 | -1.0811 | 0.0000 | |
| GSE53624 | KCND3 | 3752 | 96868 | -0.9863 | 0.0000 | |
| GSE63941 | KCND3 | 3752 | 211301_at | -0.1271 | 0.3531 | |
| GSE77861 | KCND3 | 3752 | 211827_s_at | -0.0714 | 0.5336 | |
| GSE97050 | KCND3 | 3752 | A_23_P692 | -0.8004 | 0.3104 | |
| SRP133303 | KCND3 | 3752 | RNAseq | -1.3219 | 0.0002 | |
| SRP159526 | KCND3 | 3752 | RNAseq | -1.1256 | 0.1002 | |
| SRP193095 | KCND3 | 3752 | RNAseq | 0.1651 | 0.5944 | |
| TCGA | KCND3 | 3752 | RNAseq | -0.8048 | 0.0068 |
Upregulated datasets: 0; Downregulated datasets: 3.
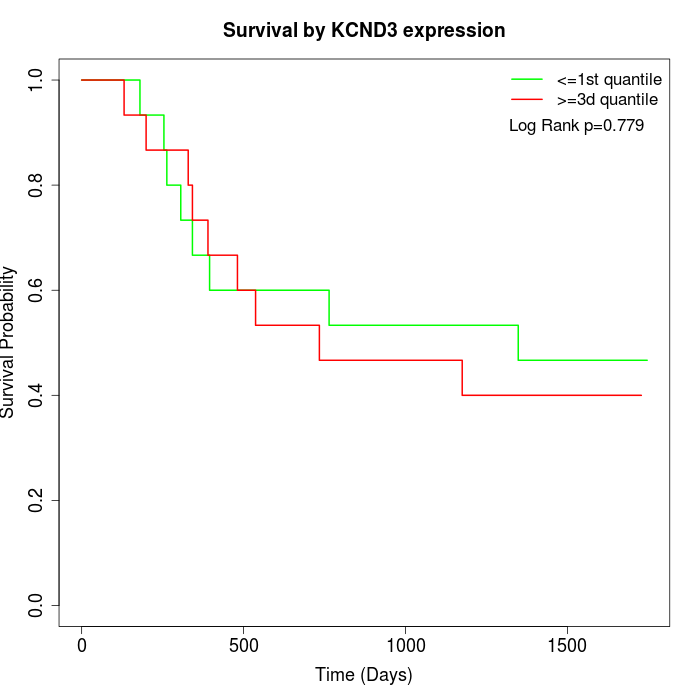
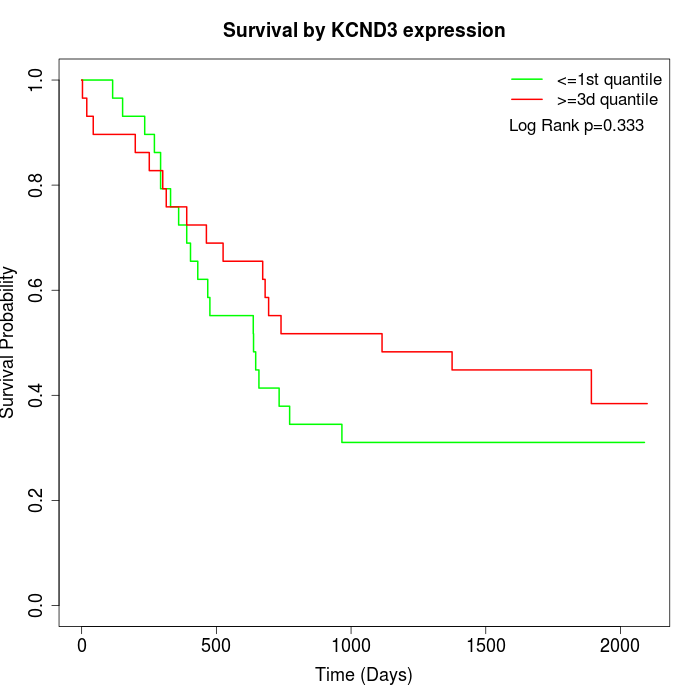
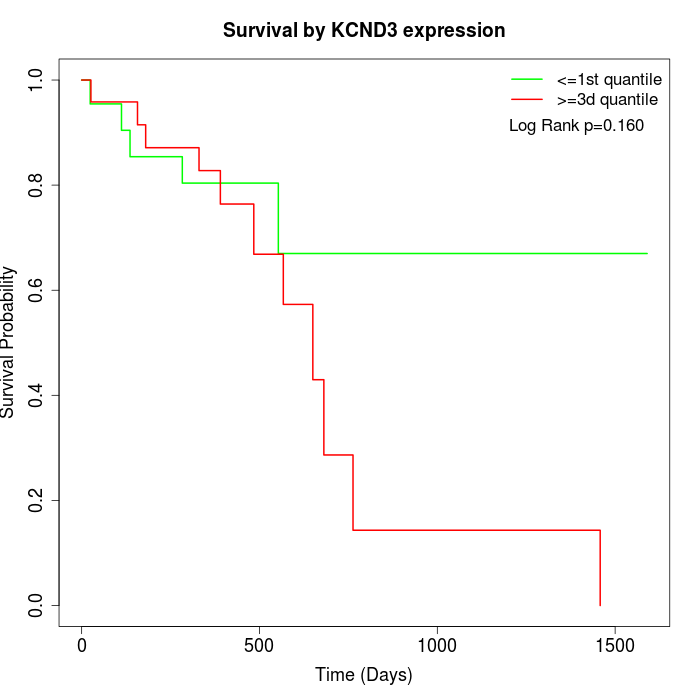
Survival by KCND3 expression:
Note: Click image to view full size file.
Copy number change of KCND3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCND3 | 3752 | 0 | 10 | 20 | |
| GSE20123 | KCND3 | 3752 | 0 | 10 | 20 | |
| GSE43470 | KCND3 | 3752 | 0 | 8 | 35 | |
| GSE46452 | KCND3 | 3752 | 2 | 1 | 56 | |
| GSE47630 | KCND3 | 3752 | 9 | 5 | 26 | |
| GSE54993 | KCND3 | 3752 | 0 | 1 | 69 | |
| GSE54994 | KCND3 | 3752 | 7 | 4 | 42 | |
| GSE60625 | KCND3 | 3752 | 0 | 0 | 11 | |
| GSE74703 | KCND3 | 3752 | 0 | 7 | 29 | |
| GSE74704 | KCND3 | 3752 | 0 | 6 | 14 | |
| TCGA | KCND3 | 3752 | 7 | 27 | 62 |
Total number of gains: 25; Total number of losses: 79; Total Number of normals: 384.
Somatic mutations of KCND3:
Generating mutation plots.
Highly correlated genes for KCND3:
Showing top 20/980 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCND3 | FOXF1 | 0.803544 | 4 | 0 | 4 |
| KCND3 | PSKH1 | 0.798343 | 3 | 0 | 3 |
| KCND3 | C16orf86 | 0.786855 | 3 | 0 | 3 |
| KCND3 | BEND5 | 0.777994 | 3 | 0 | 3 |
| KCND3 | AKT3 | 0.772607 | 3 | 0 | 3 |
| KCND3 | EBF1 | 0.769296 | 3 | 0 | 3 |
| KCND3 | FRZB | 0.76051 | 3 | 0 | 3 |
| KCND3 | STAC | 0.756048 | 3 | 0 | 3 |
| KCND3 | UBXN10 | 0.753784 | 4 | 0 | 4 |
| KCND3 | PLCB4 | 0.751865 | 3 | 0 | 3 |
| KCND3 | CACNA1H | 0.749671 | 5 | 0 | 4 |
| KCND3 | RILP | 0.747659 | 3 | 0 | 3 |
| KCND3 | TAB1 | 0.742688 | 3 | 0 | 3 |
| KCND3 | RUNX1T1 | 0.739729 | 7 | 0 | 7 |
| KCND3 | MSRB3 | 0.735809 | 4 | 0 | 4 |
| KCND3 | PI16 | 0.735517 | 5 | 0 | 5 |
| KCND3 | NES | 0.733252 | 3 | 0 | 3 |
| KCND3 | TCF7L1 | 0.733074 | 3 | 0 | 3 |
| KCND3 | CASQ2 | 0.732994 | 6 | 0 | 5 |
| KCND3 | SORCS1 | 0.730603 | 5 | 0 | 5 |
For details and further investigation, click here