| Full name: potassium calcium-activated channel subfamily N member 1 | Alias Symbol: KCa2.1|hSK1 | ||
| Type: protein-coding gene | Cytoband: 19p13.11 | ||
| Entrez ID: 3780 | HGNC ID: HGNC:6290 | Ensembl Gene: ENSG00000105642 | OMIM ID: 602982 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNN1 | 3780 | 206231_at | -0.0088 | 0.9862 | |
| GSE20347 | KCNN1 | 3780 | 206231_at | 0.0181 | 0.8754 | |
| GSE23400 | KCNN1 | 3780 | 206231_at | -0.1071 | 0.0093 | |
| GSE26886 | KCNN1 | 3780 | 206231_at | 0.1508 | 0.2600 | |
| GSE29001 | KCNN1 | 3780 | 206231_at | 0.0407 | 0.8198 | |
| GSE38129 | KCNN1 | 3780 | 206231_at | -0.0359 | 0.6886 | |
| GSE45670 | KCNN1 | 3780 | 206231_at | 0.1281 | 0.1036 | |
| GSE53622 | KCNN1 | 3780 | 39909 | -0.0290 | 0.8304 | |
| GSE53624 | KCNN1 | 3780 | 39909 | -0.3366 | 0.0009 | |
| GSE63941 | KCNN1 | 3780 | 206231_at | 0.2535 | 0.1634 | |
| GSE77861 | KCNN1 | 3780 | 206231_at | 0.0010 | 0.9958 | |
| GSE97050 | KCNN1 | 3780 | A_23_P119573 | 0.1388 | 0.5016 | |
| TCGA | KCNN1 | 3780 | RNAseq | 0.6573 | 0.1863 |
Upregulated datasets: 0; Downregulated datasets: 0.
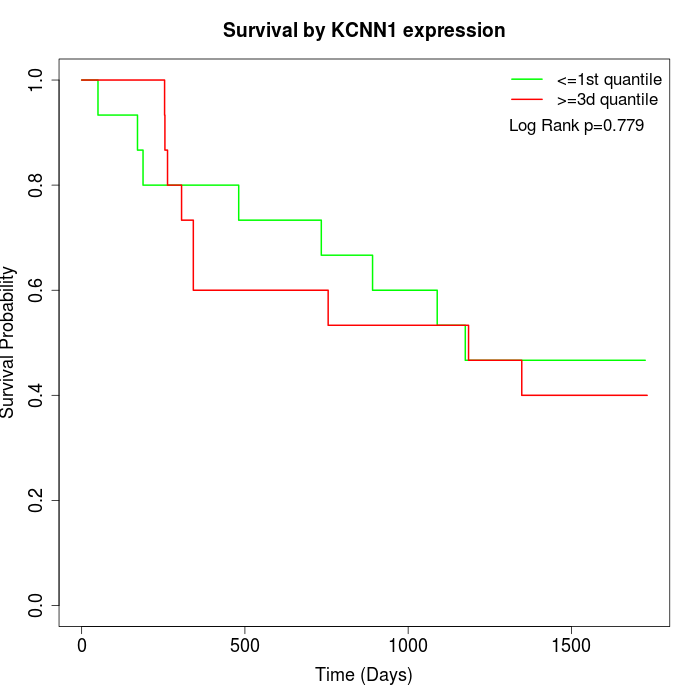
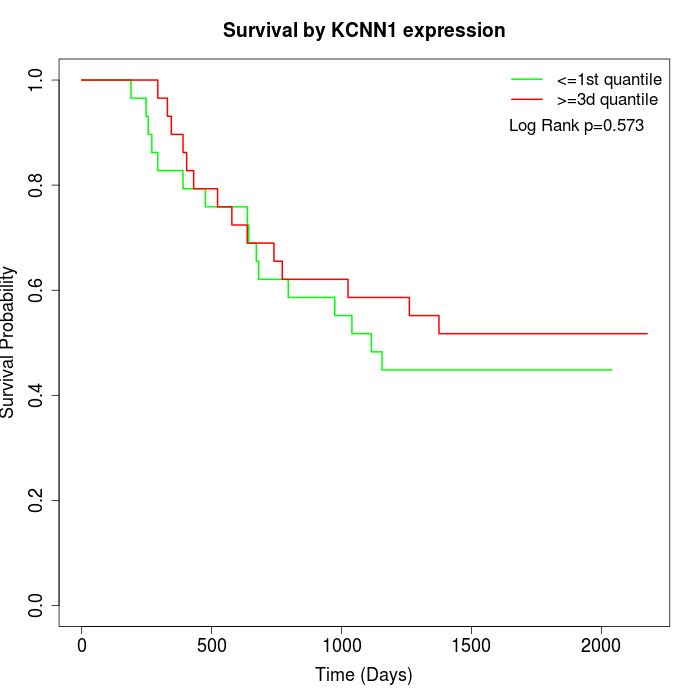
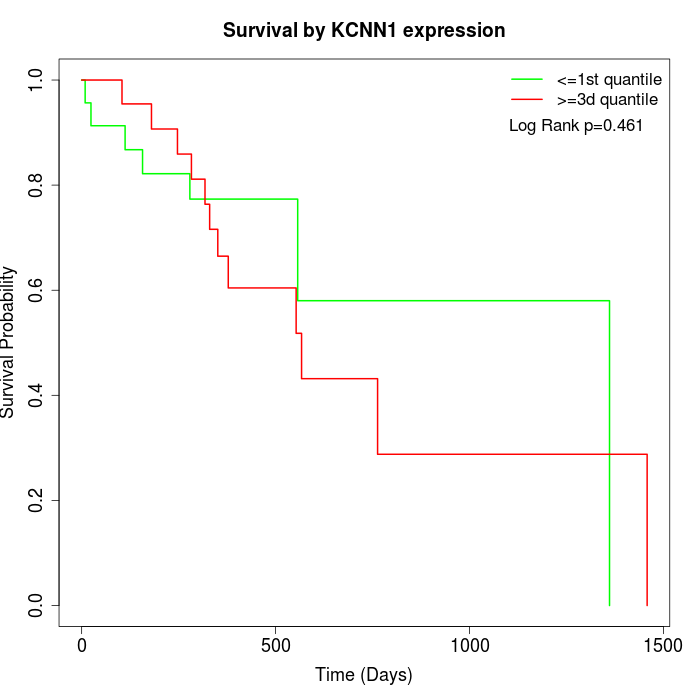
Survival by KCNN1 expression:
Note: Click image to view full size file.
Copy number change of KCNN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNN1 | 3780 | 4 | 4 | 22 | |
| GSE20123 | KCNN1 | 3780 | 3 | 2 | 25 | |
| GSE43470 | KCNN1 | 3780 | 2 | 6 | 35 | |
| GSE46452 | KCNN1 | 3780 | 47 | 1 | 11 | |
| GSE47630 | KCNN1 | 3780 | 4 | 8 | 28 | |
| GSE54993 | KCNN1 | 3780 | 15 | 4 | 51 | |
| GSE54994 | KCNN1 | 3780 | 6 | 14 | 33 | |
| GSE60625 | KCNN1 | 3780 | 9 | 0 | 2 | |
| GSE74703 | KCNN1 | 3780 | 2 | 4 | 30 | |
| GSE74704 | KCNN1 | 3780 | 0 | 2 | 18 |
Total number of gains: 92; Total number of losses: 45; Total Number of normals: 255.
Somatic mutations of KCNN1:
Generating mutation plots.
Highly correlated genes for KCNN1:
Showing top 20/829 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNN1 | DERL3 | 0.762935 | 3 | 0 | 3 |
| KCNN1 | LMAN1 | 0.743125 | 3 | 0 | 3 |
| KCNN1 | FPGS | 0.732089 | 3 | 0 | 3 |
| KCNN1 | KHDC1 | 0.714717 | 3 | 0 | 3 |
| KCNN1 | CHML | 0.712789 | 3 | 0 | 3 |
| KCNN1 | LIPF | 0.709149 | 4 | 0 | 4 |
| KCNN1 | IQSEC1 | 0.708733 | 3 | 0 | 3 |
| KCNN1 | NTNG1 | 0.699039 | 4 | 0 | 4 |
| KCNN1 | PHACTR1 | 0.691612 | 4 | 0 | 4 |
| KCNN1 | RPGRIP1 | 0.69154 | 6 | 0 | 6 |
| KCNN1 | PON1 | 0.691244 | 4 | 0 | 4 |
| KCNN1 | RIT2 | 0.687854 | 4 | 0 | 3 |
| KCNN1 | SNAI3 | 0.683805 | 3 | 0 | 3 |
| KCNN1 | GUCA2B | 0.683493 | 6 | 0 | 6 |
| KCNN1 | SIT1 | 0.681641 | 4 | 0 | 4 |
| KCNN1 | EHMT2 | 0.681351 | 4 | 0 | 4 |
| KCNN1 | CCDC7 | 0.679545 | 5 | 0 | 5 |
| KCNN1 | NEU3 | 0.678034 | 6 | 0 | 5 |
| KCNN1 | AMELY | 0.676882 | 4 | 0 | 4 |
| KCNN1 | MEIS3 | 0.676314 | 4 | 0 | 4 |
For details and further investigation, click here