| Full name: HCLS1 associated protein X-1 | Alias Symbol: HS1BP1|HCLSBP1 | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 10456 | HGNC ID: HGNC:16915 | Ensembl Gene: ENSG00000143575 | OMIM ID: 605998 |
| Drug and gene relationship at DGIdb | |||
Expression of HAX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HAX1 | 10456 | 201145_at | 0.2606 | 0.1401 | |
| GSE20347 | HAX1 | 10456 | 201145_at | 0.2454 | 0.0917 | |
| GSE23400 | HAX1 | 10456 | 201145_at | 0.1590 | 0.0134 | |
| GSE26886 | HAX1 | 10456 | 201145_at | 0.3936 | 0.0368 | |
| GSE29001 | HAX1 | 10456 | 201145_at | 0.2056 | 0.6532 | |
| GSE38129 | HAX1 | 10456 | 201145_at | 0.2603 | 0.0055 | |
| GSE45670 | HAX1 | 10456 | 201145_at | 0.2005 | 0.1156 | |
| GSE53622 | HAX1 | 10456 | 17169 | 0.2623 | 0.0000 | |
| GSE53624 | HAX1 | 10456 | 17169 | 0.3818 | 0.0000 | |
| GSE63941 | HAX1 | 10456 | 201145_at | 1.0502 | 0.0185 | |
| GSE77861 | HAX1 | 10456 | 201145_at | 0.3088 | 0.0530 | |
| GSE97050 | HAX1 | 10456 | A_23_P115223 | 0.0500 | 0.9318 | |
| SRP007169 | HAX1 | 10456 | RNAseq | -0.4906 | 0.0865 | |
| SRP008496 | HAX1 | 10456 | RNAseq | -0.2265 | 0.2677 | |
| SRP064894 | HAX1 | 10456 | RNAseq | 0.1435 | 0.5382 | |
| SRP133303 | HAX1 | 10456 | RNAseq | 0.2774 | 0.0378 | |
| SRP159526 | HAX1 | 10456 | RNAseq | 0.4814 | 0.0001 | |
| SRP193095 | HAX1 | 10456 | RNAseq | -0.0590 | 0.6199 | |
| SRP219564 | HAX1 | 10456 | RNAseq | 0.2467 | 0.4146 | |
| TCGA | HAX1 | 10456 | RNAseq | 0.0153 | 0.7470 |
Upregulated datasets: 1; Downregulated datasets: 0.
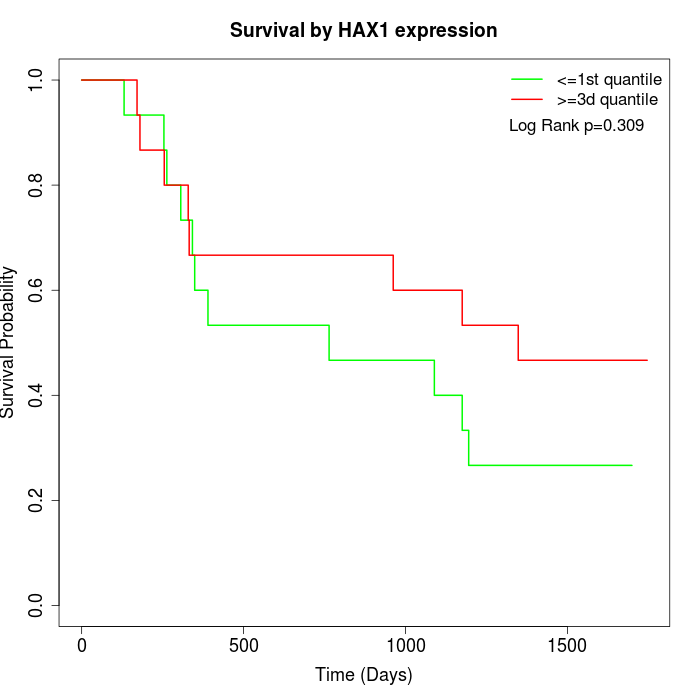
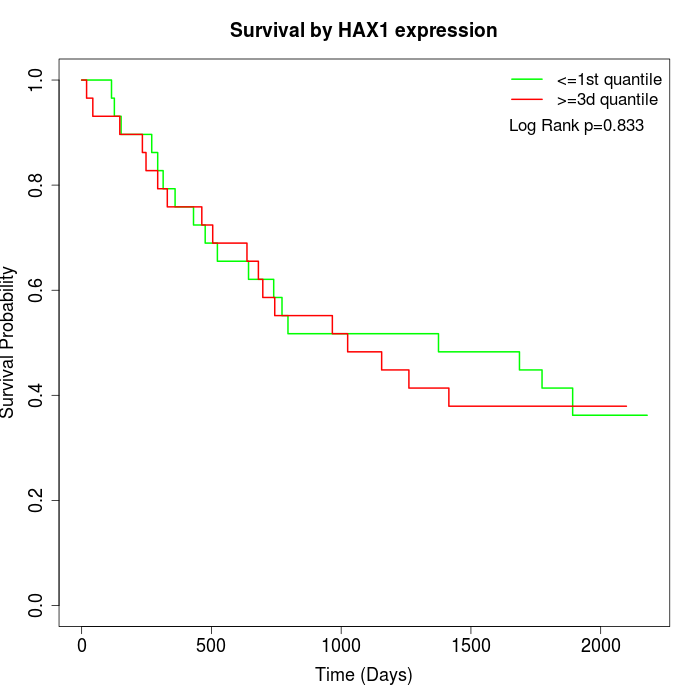
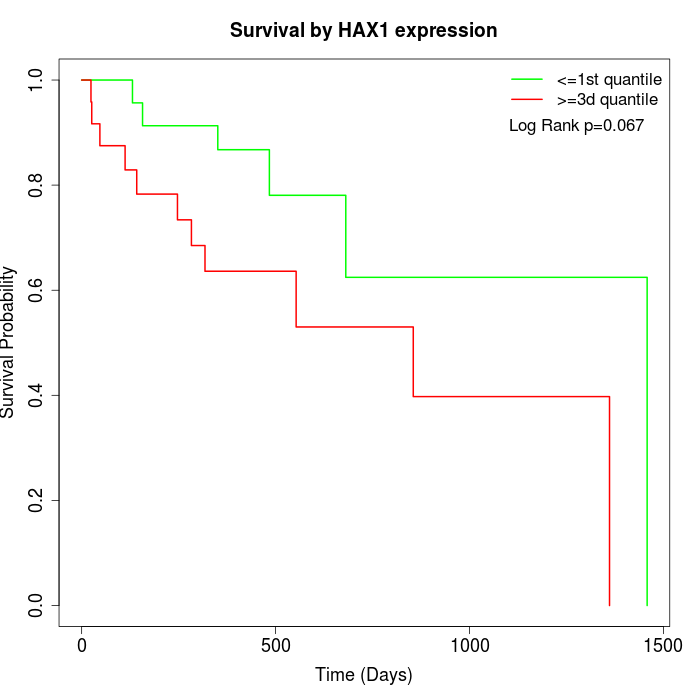
Survival by HAX1 expression:
Note: Click image to view full size file.
Copy number change of HAX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HAX1 | 10456 | 14 | 0 | 16 | |
| GSE20123 | HAX1 | 10456 | 14 | 0 | 16 | |
| GSE43470 | HAX1 | 10456 | 7 | 2 | 34 | |
| GSE46452 | HAX1 | 10456 | 2 | 1 | 56 | |
| GSE47630 | HAX1 | 10456 | 14 | 0 | 26 | |
| GSE54993 | HAX1 | 10456 | 0 | 4 | 66 | |
| GSE54994 | HAX1 | 10456 | 16 | 0 | 37 | |
| GSE60625 | HAX1 | 10456 | 0 | 0 | 11 | |
| GSE74703 | HAX1 | 10456 | 6 | 1 | 29 | |
| GSE74704 | HAX1 | 10456 | 7 | 0 | 13 | |
| TCGA | HAX1 | 10456 | 38 | 2 | 56 |
Total number of gains: 118; Total number of losses: 10; Total Number of normals: 360.
Somatic mutations of HAX1:
Generating mutation plots.
Highly correlated genes for HAX1:
Showing top 20/1153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HAX1 | RLIM | 0.807703 | 3 | 0 | 3 |
| HAX1 | HMG20A | 0.800574 | 3 | 0 | 3 |
| HAX1 | AZIN1 | 0.795694 | 3 | 0 | 3 |
| HAX1 | KTI12 | 0.761156 | 3 | 0 | 3 |
| HAX1 | RNF4 | 0.737735 | 3 | 0 | 3 |
| HAX1 | FANCL | 0.727238 | 3 | 0 | 3 |
| HAX1 | HEXA | 0.725472 | 3 | 0 | 3 |
| HAX1 | PITX2 | 0.723089 | 3 | 0 | 3 |
| HAX1 | BLCAP | 0.717824 | 3 | 0 | 3 |
| HAX1 | SLC35B2 | 0.71523 | 3 | 0 | 3 |
| HAX1 | DGKH | 0.715129 | 3 | 0 | 3 |
| HAX1 | EARS2 | 0.709749 | 3 | 0 | 3 |
| HAX1 | CERCAM | 0.705964 | 3 | 0 | 3 |
| HAX1 | CEP250 | 0.705013 | 4 | 0 | 3 |
| HAX1 | ZNF584 | 0.702721 | 3 | 0 | 3 |
| HAX1 | PIEZO1 | 0.700977 | 4 | 0 | 3 |
| HAX1 | BTN3A2 | 0.69624 | 3 | 0 | 3 |
| HAX1 | GOLGA5 | 0.696191 | 3 | 0 | 3 |
| HAX1 | PTPMT1 | 0.694114 | 5 | 0 | 5 |
| HAX1 | TMEM167A | 0.693951 | 3 | 0 | 3 |
For details and further investigation, click here