| Full name: KTI12 chromatin associated homolog | Alias Symbol: TOT4|MGC20419|SBBI81 | ||
| Type: protein-coding gene | Cytoband: 1p32.3 | ||
| Entrez ID: 112970 | HGNC ID: HGNC:25160 | Ensembl Gene: ENSG00000198841 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of KTI12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | KTI12 | 112970 | 130400 | 0.4860 | 0.0000 | |
| GSE53624 | KTI12 | 112970 | 130400 | 0.7119 | 0.0000 | |
| GSE97050 | KTI12 | 112970 | A_23_P103276 | 0.3903 | 0.2018 | |
| SRP007169 | KTI12 | 112970 | RNAseq | 0.4423 | 0.3700 | |
| SRP008496 | KTI12 | 112970 | RNAseq | 0.6617 | 0.0644 | |
| SRP064894 | KTI12 | 112970 | RNAseq | 0.5779 | 0.0994 | |
| SRP159526 | KTI12 | 112970 | RNAseq | -0.3716 | 0.5463 | |
| SRP219564 | KTI12 | 112970 | RNAseq | 0.5632 | 0.1257 | |
| TCGA | KTI12 | 112970 | RNAseq | 0.0591 | 0.3673 |
Upregulated datasets: 0; Downregulated datasets: 0.
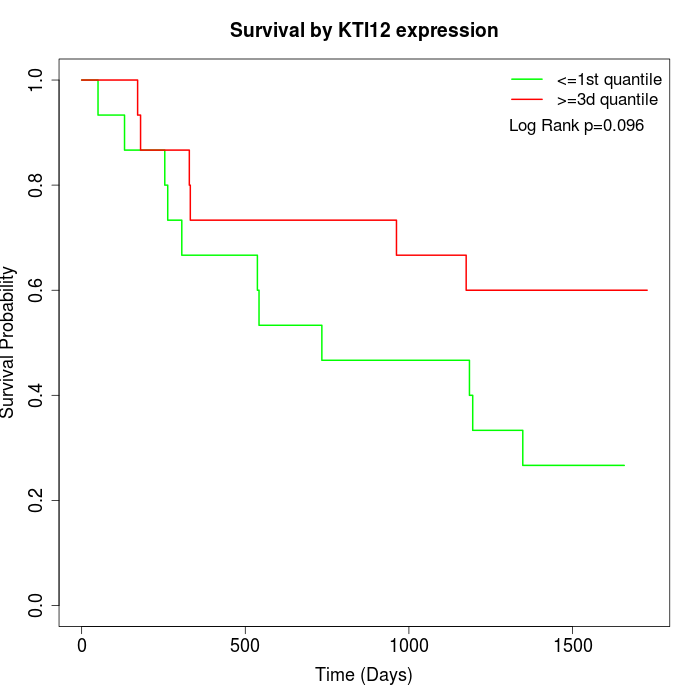
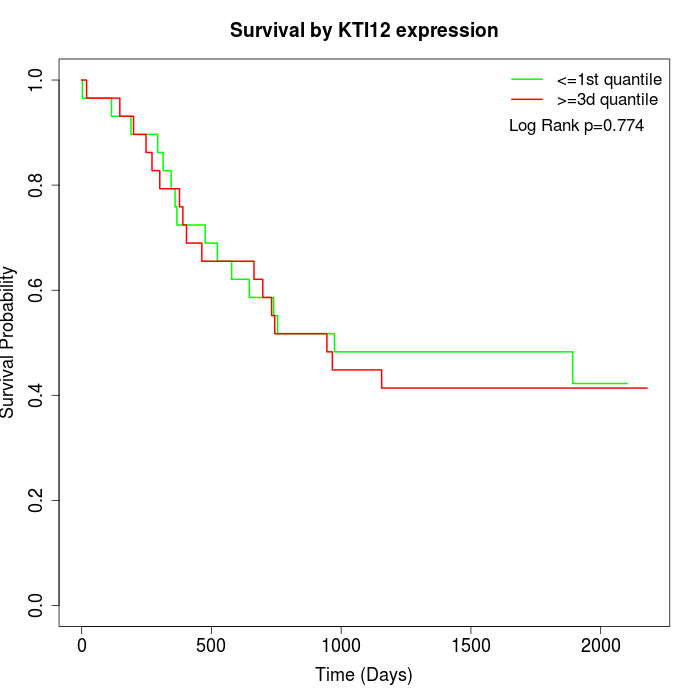
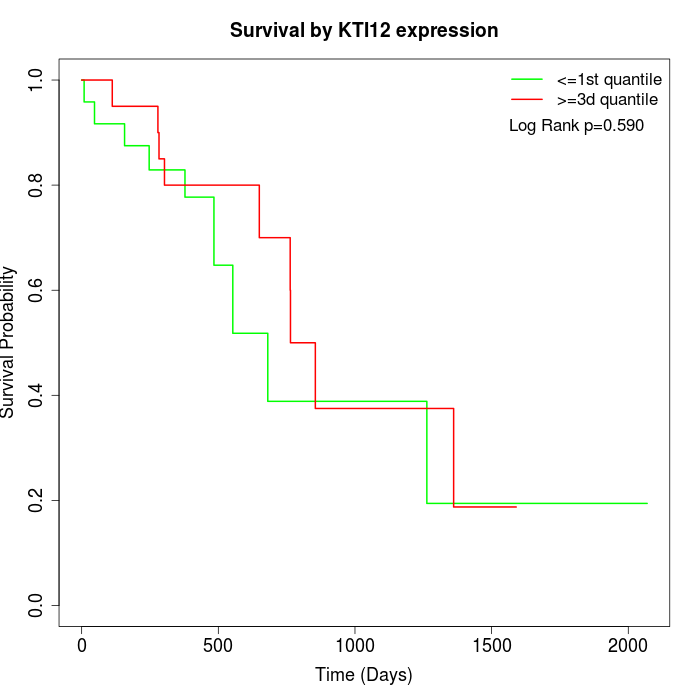
Survival by KTI12 expression:
Note: Click image to view full size file.
Copy number change of KTI12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KTI12 | 112970 | 3 | 4 | 23 | |
| GSE20123 | KTI12 | 112970 | 3 | 3 | 24 | |
| GSE43470 | KTI12 | 112970 | 5 | 4 | 34 | |
| GSE46452 | KTI12 | 112970 | 2 | 1 | 56 | |
| GSE47630 | KTI12 | 112970 | 8 | 4 | 28 | |
| GSE54993 | KTI12 | 112970 | 0 | 1 | 69 | |
| GSE54994 | KTI12 | 112970 | 11 | 2 | 40 | |
| GSE60625 | KTI12 | 112970 | 0 | 0 | 11 | |
| GSE74703 | KTI12 | 112970 | 4 | 2 | 30 | |
| GSE74704 | KTI12 | 112970 | 2 | 0 | 18 | |
| TCGA | KTI12 | 112970 | 10 | 18 | 68 |
Total number of gains: 48; Total number of losses: 39; Total Number of normals: 401.
Somatic mutations of KTI12:
Generating mutation plots.
Highly correlated genes for KTI12:
Showing top 20/479 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KTI12 | MRPL37 | 0.822456 | 3 | 0 | 3 |
| KTI12 | FARSA | 0.800167 | 3 | 0 | 3 |
| KTI12 | CLN6 | 0.799186 | 3 | 0 | 3 |
| KTI12 | MRPS34 | 0.795572 | 3 | 0 | 3 |
| KTI12 | ZC3HC1 | 0.791274 | 3 | 0 | 3 |
| KTI12 | WDR4 | 0.783589 | 3 | 0 | 3 |
| KTI12 | GRWD1 | 0.783007 | 3 | 0 | 3 |
| KTI12 | NCF2 | 0.781836 | 3 | 0 | 3 |
| KTI12 | ATP6V0B | 0.780422 | 3 | 0 | 3 |
| KTI12 | SMG8 | 0.780329 | 3 | 0 | 3 |
| KTI12 | METTL23 | 0.776957 | 3 | 0 | 3 |
| KTI12 | ARPC1B | 0.774857 | 3 | 0 | 3 |
| KTI12 | RNF34 | 0.774805 | 3 | 0 | 3 |
| KTI12 | YIF1A | 0.774501 | 3 | 0 | 3 |
| KTI12 | SLC25A32 | 0.77193 | 3 | 0 | 3 |
| KTI12 | ZNF317 | 0.771777 | 3 | 0 | 3 |
| KTI12 | ADRM1 | 0.770494 | 3 | 0 | 3 |
| KTI12 | DHX33 | 0.769699 | 3 | 0 | 3 |
| KTI12 | MRPL2 | 0.76939 | 3 | 0 | 3 |
| KTI12 | CLSPN | 0.768327 | 3 | 0 | 3 |
For details and further investigation, click here