| Full name: holocytochrome c synthase | Alias Symbol: CCHL | ||
| Type: protein-coding gene | Cytoband: Xp22.2 | ||
| Entrez ID: 3052 | HGNC ID: HGNC:4837 | Ensembl Gene: ENSG00000004961 | OMIM ID: 300056 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of HCCS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCCS | 3052 | 203746_s_at | -0.0451 | 0.9204 | |
| GSE20347 | HCCS | 3052 | 203746_s_at | -0.1815 | 0.4512 | |
| GSE23400 | HCCS | 3052 | 203746_s_at | 0.0136 | 0.8723 | |
| GSE26886 | HCCS | 3052 | 203746_s_at | -0.5805 | 0.0192 | |
| GSE29001 | HCCS | 3052 | 203746_s_at | -0.0339 | 0.9329 | |
| GSE38129 | HCCS | 3052 | 203746_s_at | -0.0809 | 0.6750 | |
| GSE45670 | HCCS | 3052 | 203746_s_at | 0.3303 | 0.0443 | |
| GSE53622 | HCCS | 3052 | 88132 | -0.2275 | 0.0047 | |
| GSE53624 | HCCS | 3052 | 88132 | -0.0400 | 0.7466 | |
| GSE63941 | HCCS | 3052 | 203746_s_at | 0.4930 | 0.3486 | |
| GSE77861 | HCCS | 3052 | 203746_s_at | -0.1375 | 0.6551 | |
| GSE97050 | HCCS | 3052 | A_23_P257945 | -0.0923 | 0.7400 | |
| SRP007169 | HCCS | 3052 | RNAseq | 0.2277 | 0.7338 | |
| SRP064894 | HCCS | 3052 | RNAseq | 0.0110 | 0.9483 | |
| SRP133303 | HCCS | 3052 | RNAseq | 0.3968 | 0.0721 | |
| SRP159526 | HCCS | 3052 | RNAseq | -0.0120 | 0.9697 | |
| SRP193095 | HCCS | 3052 | RNAseq | -0.1461 | 0.2059 | |
| SRP219564 | HCCS | 3052 | RNAseq | 0.0990 | 0.8107 | |
| TCGA | HCCS | 3052 | RNAseq | -0.0440 | 0.4399 |
Upregulated datasets: 0; Downregulated datasets: 0.
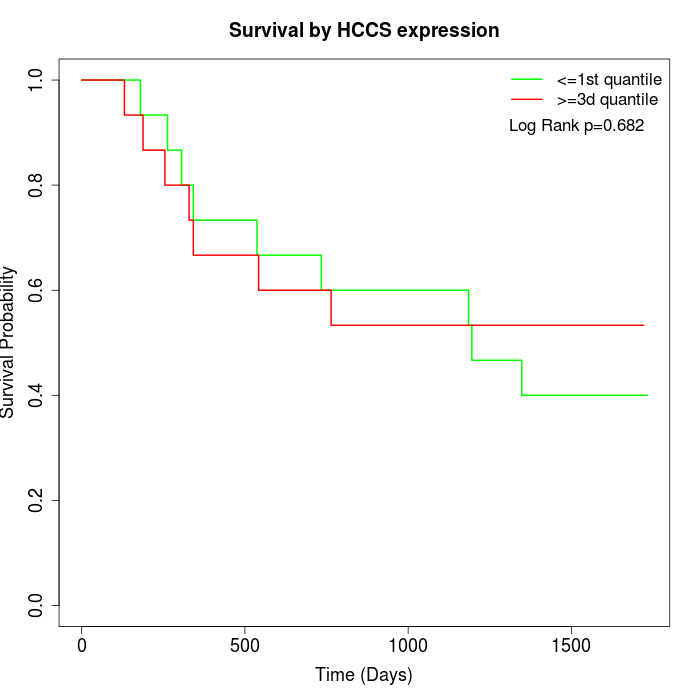
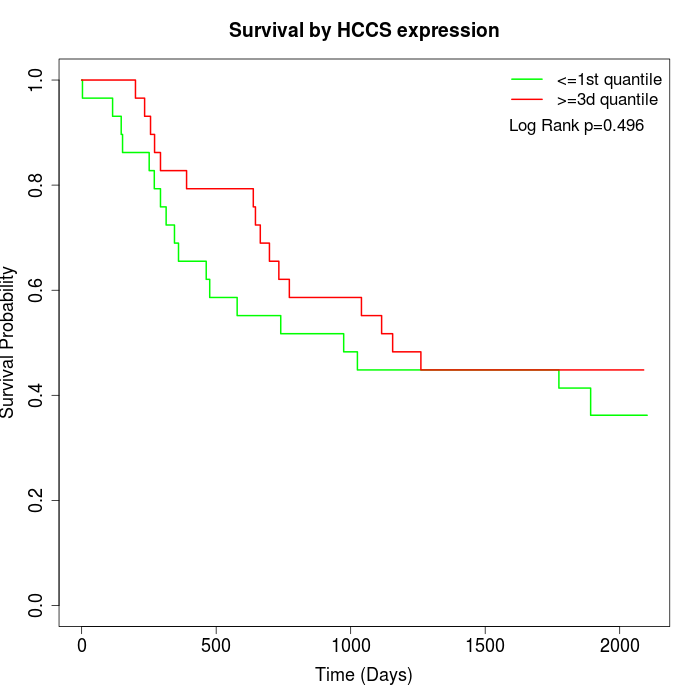
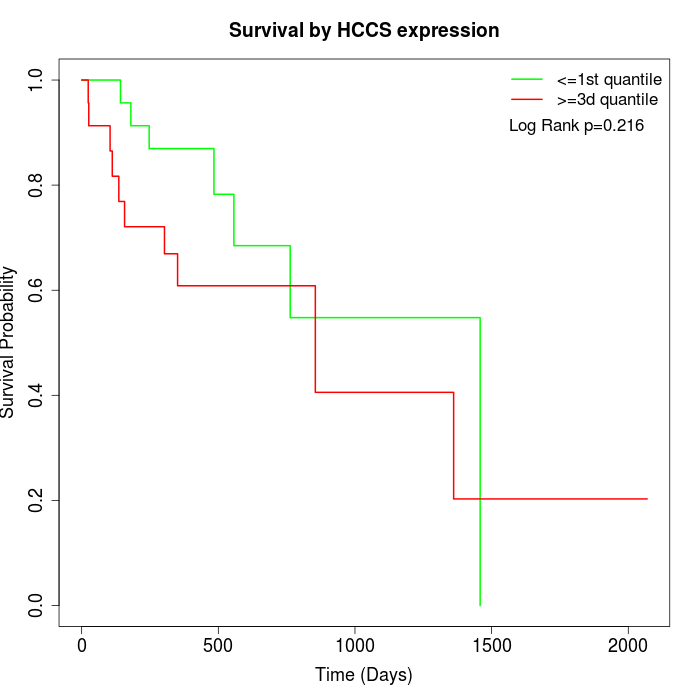
Survival by HCCS expression:
Note: Click image to view full size file.
Copy number change of HCCS:
No record found for this gene.
Somatic mutations of HCCS:
Generating mutation plots.
Highly correlated genes for HCCS:
Showing top 20/447 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCCS | MRRF | 0.79363 | 3 | 0 | 3 |
| HCCS | GNL3 | 0.792096 | 3 | 0 | 3 |
| HCCS | SUPT4H1 | 0.783961 | 3 | 0 | 3 |
| HCCS | HBS1L | 0.78076 | 3 | 0 | 3 |
| HCCS | SDHAF2 | 0.779945 | 3 | 0 | 3 |
| HCCS | TYW3 | 0.776154 | 3 | 0 | 3 |
| HCCS | C3orf38 | 0.773764 | 3 | 0 | 3 |
| HCCS | CNOT10 | 0.772733 | 3 | 0 | 3 |
| HCCS | FAM32A | 0.77134 | 3 | 0 | 3 |
| HCCS | LARP1B | 0.767758 | 3 | 0 | 3 |
| HCCS | RPGR | 0.765572 | 3 | 0 | 3 |
| HCCS | MRPL1 | 0.760536 | 3 | 0 | 3 |
| HCCS | SNX12 | 0.758919 | 3 | 0 | 3 |
| HCCS | TMEM53 | 0.747715 | 3 | 0 | 3 |
| HCCS | SNAPIN | 0.741939 | 3 | 0 | 3 |
| HCCS | DUSP22 | 0.741597 | 3 | 0 | 3 |
| HCCS | KANSL2 | 0.74112 | 4 | 0 | 4 |
| HCCS | SAP30L | 0.736302 | 3 | 0 | 3 |
| HCCS | RHBDD1 | 0.733449 | 3 | 0 | 3 |
| HCCS | TUFM | 0.729603 | 3 | 0 | 3 |
For details and further investigation, click here