| Full name: dual specificity phosphatase 22 | Alias Symbol: MKPX|JSP1|JKAP|VHX | ||
| Type: protein-coding gene | Cytoband: 6p25.3 | ||
| Entrez ID: 56940 | HGNC ID: HGNC:16077 | Ensembl Gene: ENSG00000112679 | OMIM ID: 616778 |
| Drug and gene relationship at DGIdb | |||
Expression of DUSP22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP22 | 56940 | 218845_at | -0.7662 | 0.1088 | |
| GSE20347 | DUSP22 | 56940 | 218845_at | -1.2556 | 0.0000 | |
| GSE23400 | DUSP22 | 56940 | 218845_at | -0.5457 | 0.0000 | |
| GSE26886 | DUSP22 | 56940 | 218845_at | -0.9031 | 0.0000 | |
| GSE29001 | DUSP22 | 56940 | 218845_at | -0.6996 | 0.0154 | |
| GSE38129 | DUSP22 | 56940 | 218845_at | -0.9858 | 0.0000 | |
| GSE45670 | DUSP22 | 56940 | 218845_at | -0.3295 | 0.1100 | |
| GSE53622 | DUSP22 | 56940 | 128651 | -1.1186 | 0.0000 | |
| GSE53624 | DUSP22 | 56940 | 128651 | -1.2863 | 0.0000 | |
| GSE63941 | DUSP22 | 56940 | 218845_at | -0.4665 | 0.2164 | |
| GSE77861 | DUSP22 | 56940 | 218845_at | -0.9472 | 0.0020 | |
| GSE97050 | DUSP22 | 56940 | A_23_P7896 | -0.4308 | 0.1782 | |
| SRP007169 | DUSP22 | 56940 | RNAseq | -1.5889 | 0.0006 | |
| SRP008496 | DUSP22 | 56940 | RNAseq | -1.7088 | 0.0000 | |
| SRP064894 | DUSP22 | 56940 | RNAseq | -0.4841 | 0.0469 | |
| SRP133303 | DUSP22 | 56940 | RNAseq | -0.8268 | 0.0000 | |
| SRP159526 | DUSP22 | 56940 | RNAseq | -0.7431 | 0.0497 | |
| SRP193095 | DUSP22 | 56940 | RNAseq | -0.8727 | 0.0000 | |
| SRP219564 | DUSP22 | 56940 | RNAseq | -0.5667 | 0.2396 | |
| TCGA | DUSP22 | 56940 | RNAseq | -0.2745 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 5.
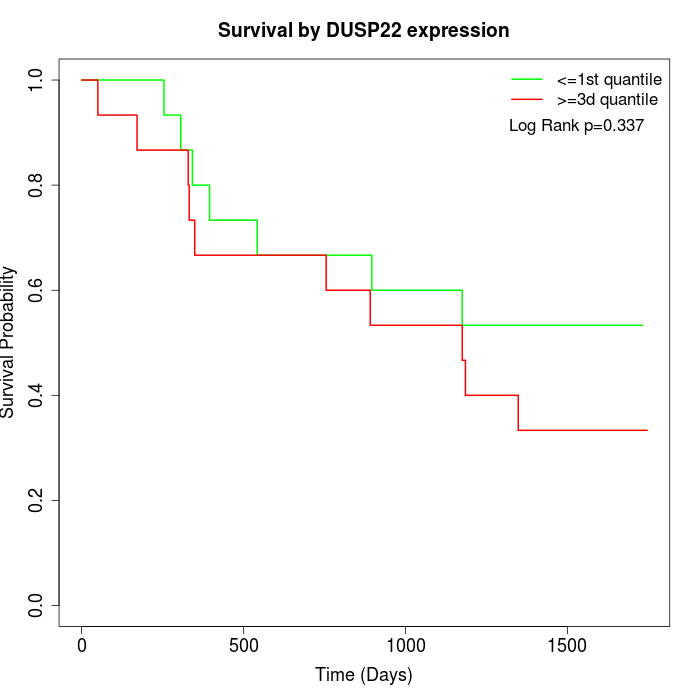
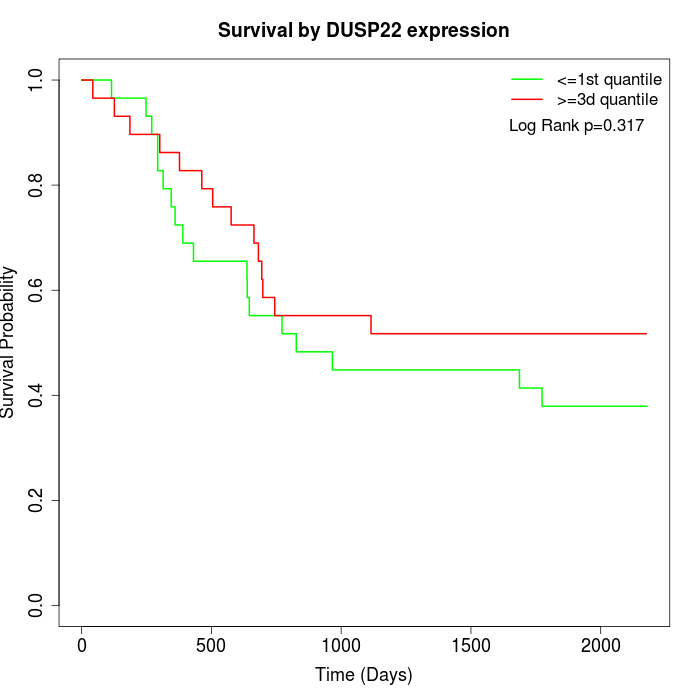
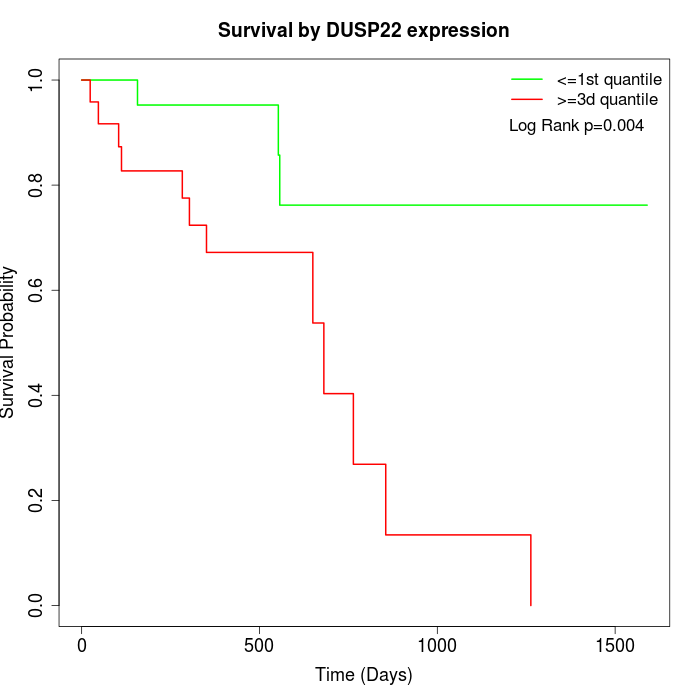
Survival by DUSP22 expression:
Note: Click image to view full size file.
Copy number change of DUSP22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP22 | 56940 | 3 | 5 | 22 | |
| GSE20123 | DUSP22 | 56940 | 3 | 5 | 22 | |
| GSE43470 | DUSP22 | 56940 | 8 | 4 | 31 | |
| GSE46452 | DUSP22 | 56940 | 1 | 10 | 48 | |
| GSE47630 | DUSP22 | 56940 | 7 | 9 | 24 | |
| GSE54993 | DUSP22 | 56940 | 1 | 3 | 66 | |
| GSE54994 | DUSP22 | 56940 | 9 | 2 | 42 | |
| GSE60625 | DUSP22 | 56940 | 0 | 1 | 10 | |
| GSE74703 | DUSP22 | 56940 | 7 | 3 | 26 | |
| GSE74704 | DUSP22 | 56940 | 2 | 2 | 16 | |
| TCGA | DUSP22 | 56940 | 16 | 29 | 51 |
Total number of gains: 57; Total number of losses: 73; Total Number of normals: 358.
Somatic mutations of DUSP22:
Generating mutation plots.
Highly correlated genes for DUSP22:
Showing top 20/1794 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP22 | ALKBH7 | 0.794337 | 5 | 0 | 5 |
| DUSP22 | ZCCHC9 | 0.785411 | 3 | 0 | 3 |
| DUSP22 | FAM174A | 0.775387 | 6 | 0 | 6 |
| DUSP22 | TBC1D22B | 0.77528 | 5 | 0 | 5 |
| DUSP22 | FBXL17 | 0.772263 | 5 | 0 | 5 |
| DUSP22 | TMEM120A | 0.771069 | 6 | 0 | 6 |
| DUSP22 | ACAA1 | 0.767456 | 8 | 0 | 8 |
| DUSP22 | CMPK1 | 0.764617 | 10 | 0 | 10 |
| DUSP22 | ARHGEF10L | 0.761964 | 11 | 0 | 11 |
| DUSP22 | PLEKHA7 | 0.76097 | 7 | 0 | 7 |
| DUSP22 | CCNY | 0.755276 | 3 | 0 | 3 |
| DUSP22 | CTTNBP2NL | 0.75525 | 7 | 0 | 7 |
| DUSP22 | MPP7 | 0.754781 | 7 | 0 | 7 |
| DUSP22 | GBP6 | 0.754 | 4 | 0 | 3 |
| DUSP22 | NPRL2 | 0.75033 | 5 | 0 | 5 |
| DUSP22 | BRK1 | 0.749943 | 7 | 0 | 7 |
| DUSP22 | RAB5A | 0.74702 | 11 | 0 | 11 |
| DUSP22 | FCHO2 | 0.745492 | 7 | 0 | 7 |
| DUSP22 | PITHD1 | 0.744003 | 7 | 0 | 6 |
| DUSP22 | TRNP1 | 0.743809 | 6 | 0 | 6 |
For details and further investigation, click here