| Full name: hyperpolarization activated cyclic nucleotide gated potassium channel 1 | Alias Symbol: BCNG-1|HAC-2 | ||
| Type: protein-coding gene | Cytoband: 5p12 | ||
| Entrez ID: 348980 | HGNC ID: HGNC:4845 | Ensembl Gene: ENSG00000164588 | OMIM ID: 602780 |
| Drug and gene relationship at DGIdb | |||
Expression of HCN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCN1 | 348980 | 1562563_at | -0.1013 | 0.6344 | |
| GSE26886 | HCN1 | 348980 | 1562563_at | -0.0373 | 0.8161 | |
| GSE45670 | HCN1 | 348980 | 1562563_at | -0.0757 | 0.4703 | |
| GSE53622 | HCN1 | 348980 | 24775 | -0.2445 | 0.0398 | |
| GSE53624 | HCN1 | 348980 | 24775 | -0.1702 | 0.0158 | |
| GSE63941 | HCN1 | 348980 | 1562563_at | 0.0959 | 0.5469 | |
| GSE77861 | HCN1 | 348980 | 1562563_at | -0.1361 | 0.1583 | |
| SRP133303 | HCN1 | 348980 | RNAseq | -0.0791 | 0.7521 | |
| SRP193095 | HCN1 | 348980 | RNAseq | -0.3627 | 0.0254 | |
| SRP219564 | HCN1 | 348980 | RNAseq | -0.5426 | 0.3850 | |
| TCGA | HCN1 | 348980 | RNAseq | -2.9042 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 1.
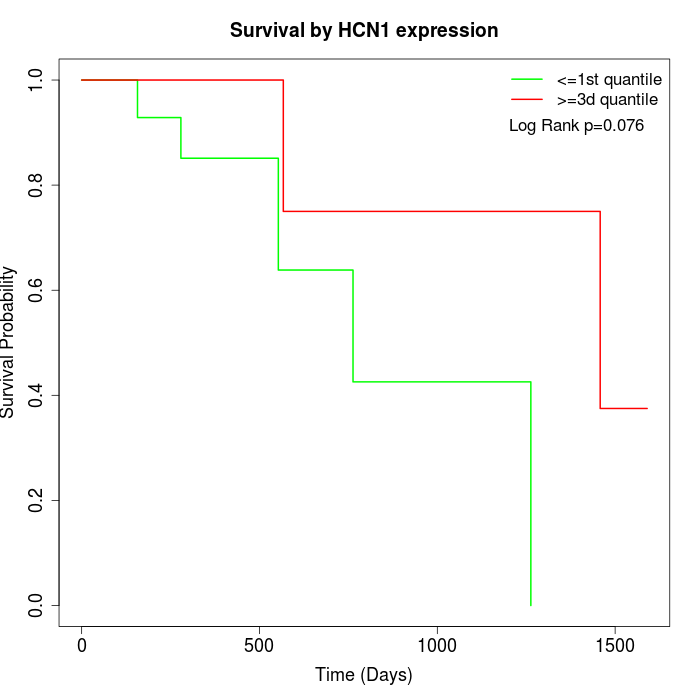
Survival by HCN1 expression:
Note: Click image to view full size file.
Copy number change of HCN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCN1 | 348980 | 9 | 1 | 20 | |
| GSE20123 | HCN1 | 348980 | 9 | 1 | 20 | |
| GSE43470 | HCN1 | 348980 | 16 | 1 | 26 | |
| GSE46452 | HCN1 | 348980 | 5 | 22 | 32 | |
| GSE47630 | HCN1 | 348980 | 6 | 12 | 22 | |
| GSE54993 | HCN1 | 348980 | 6 | 4 | 60 | |
| GSE54994 | HCN1 | 348980 | 24 | 2 | 27 | |
| GSE60625 | HCN1 | 348980 | 0 | 0 | 11 | |
| GSE74703 | HCN1 | 348980 | 13 | 1 | 22 | |
| GSE74704 | HCN1 | 348980 | 8 | 1 | 11 | |
| TCGA | HCN1 | 348980 | 51 | 20 | 25 |
Total number of gains: 147; Total number of losses: 65; Total Number of normals: 276.
Somatic mutations of HCN1:
Generating mutation plots.
Highly correlated genes for HCN1:
Showing top 20/108 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCN1 | VGLL2 | 0.71334 | 3 | 0 | 3 |
| HCN1 | KIF12 | 0.687797 | 3 | 0 | 3 |
| HCN1 | KRT73 | 0.678567 | 3 | 0 | 3 |
| HCN1 | WFDC8 | 0.67614 | 3 | 0 | 3 |
| HCN1 | TPSD1 | 0.662969 | 3 | 0 | 3 |
| HCN1 | TAL1 | 0.649305 | 4 | 0 | 4 |
| HCN1 | ALLC | 0.647229 | 3 | 0 | 3 |
| HCN1 | TSGA10IP | 0.643751 | 3 | 0 | 3 |
| HCN1 | LINC00315 | 0.64223 | 3 | 0 | 3 |
| HCN1 | HCAR3 | 0.641189 | 3 | 0 | 3 |
| HCN1 | MYL7 | 0.636405 | 3 | 0 | 3 |
| HCN1 | ANKFN1 | 0.635357 | 4 | 0 | 4 |
| HCN1 | RXFP3 | 0.635246 | 3 | 0 | 3 |
| HCN1 | F7 | 0.634704 | 3 | 0 | 3 |
| HCN1 | LINC00628 | 0.631016 | 4 | 0 | 3 |
| HCN1 | RS1 | 0.628374 | 3 | 0 | 3 |
| HCN1 | BFSP2 | 0.627855 | 3 | 0 | 3 |
| HCN1 | MIP | 0.625846 | 4 | 0 | 3 |
| HCN1 | HGFAC | 0.625246 | 3 | 0 | 3 |
| HCN1 | IFNA21 | 0.622299 | 3 | 0 | 3 |
For details and further investigation, click here