| Full name: beaded filament structural protein 2 | Alias Symbol: CP47|CP49|LIFL-L|phakinin | ||
| Type: protein-coding gene | Cytoband: 3q22.1 | ||
| Entrez ID: 8419 | HGNC ID: HGNC:1041 | Ensembl Gene: ENSG00000170819 | OMIM ID: 603212 |
| Drug and gene relationship at DGIdb | |||
Expression of BFSP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BFSP2 | 8419 | 207399_at | -0.1172 | 0.7188 | |
| GSE20347 | BFSP2 | 8419 | 207399_at | -0.1231 | 0.0751 | |
| GSE23400 | BFSP2 | 8419 | 207399_at | -0.1615 | 0.0000 | |
| GSE26886 | BFSP2 | 8419 | 207399_at | 0.0596 | 0.5654 | |
| GSE29001 | BFSP2 | 8419 | 207399_at | -0.1280 | 0.3325 | |
| GSE38129 | BFSP2 | 8419 | 207399_at | -0.0818 | 0.1864 | |
| GSE45670 | BFSP2 | 8419 | 207399_at | -0.0898 | 0.4029 | |
| GSE63941 | BFSP2 | 8419 | 207399_at | -0.0374 | 0.8420 | |
| GSE77861 | BFSP2 | 8419 | 207399_at | -0.1498 | 0.1189 | |
| TCGA | BFSP2 | 8419 | RNAseq | -0.7202 | 0.5190 |
Upregulated datasets: 0; Downregulated datasets: 0.
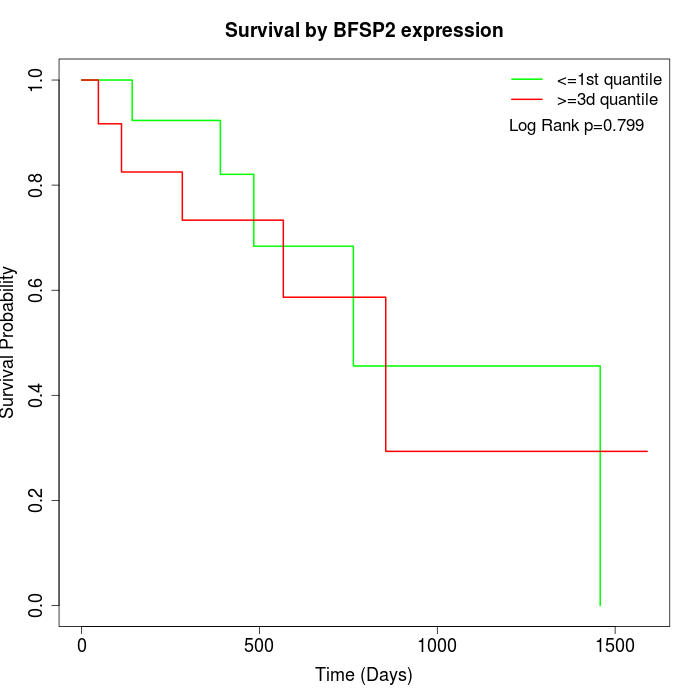
Survival by BFSP2 expression:
Note: Click image to view full size file.
Copy number change of BFSP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BFSP2 | 8419 | 16 | 0 | 14 | |
| GSE20123 | BFSP2 | 8419 | 16 | 0 | 14 | |
| GSE43470 | BFSP2 | 8419 | 21 | 0 | 22 | |
| GSE46452 | BFSP2 | 8419 | 15 | 4 | 40 | |
| GSE47630 | BFSP2 | 8419 | 18 | 3 | 19 | |
| GSE54993 | BFSP2 | 8419 | 2 | 8 | 60 | |
| GSE54994 | BFSP2 | 8419 | 33 | 2 | 18 | |
| GSE60625 | BFSP2 | 8419 | 0 | 6 | 5 | |
| GSE74703 | BFSP2 | 8419 | 18 | 0 | 18 | |
| GSE74704 | BFSP2 | 8419 | 11 | 0 | 9 | |
| TCGA | BFSP2 | 8419 | 59 | 5 | 32 |
Total number of gains: 209; Total number of losses: 28; Total Number of normals: 251.
Somatic mutations of BFSP2:
Generating mutation plots.
Highly correlated genes for BFSP2:
Showing top 20/904 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BFSP2 | LGALS13 | 0.757885 | 3 | 0 | 3 |
| BFSP2 | VGF | 0.750406 | 3 | 0 | 3 |
| BFSP2 | ARMC3 | 0.725804 | 3 | 0 | 3 |
| BFSP2 | PF4 | 0.722966 | 4 | 0 | 4 |
| BFSP2 | MVK | 0.721443 | 5 | 0 | 5 |
| BFSP2 | THRB | 0.698639 | 3 | 0 | 3 |
| BFSP2 | HBQ1 | 0.696111 | 5 | 0 | 5 |
| BFSP2 | SLC26A1 | 0.694301 | 5 | 0 | 5 |
| BFSP2 | GPR31 | 0.690912 | 5 | 0 | 5 |
| BFSP2 | COX6A2 | 0.688443 | 6 | 0 | 5 |
| BFSP2 | RIT2 | 0.68838 | 3 | 0 | 3 |
| BFSP2 | HAAO | 0.687527 | 6 | 0 | 6 |
| BFSP2 | RBP4 | 0.684885 | 3 | 0 | 3 |
| BFSP2 | KRT3 | 0.676906 | 5 | 0 | 5 |
| BFSP2 | LEFTY1 | 0.676609 | 3 | 0 | 3 |
| BFSP2 | HLA-DOA | 0.676328 | 6 | 0 | 5 |
| BFSP2 | COL19A1 | 0.674359 | 4 | 0 | 3 |
| BFSP2 | GUCA1B | 0.673262 | 4 | 0 | 4 |
| BFSP2 | INSL3 | 0.673186 | 5 | 0 | 5 |
| BFSP2 | CAPN11 | 0.67305 | 4 | 0 | 3 |
For details and further investigation, click here