| Full name: major intrinsic protein of lens fiber | Alias Symbol: MP26|LIM1|AQP0 | ||
| Type: protein-coding gene | Cytoband: 12q13 | ||
| Entrez ID: 4284 | HGNC ID: HGNC:7103 | Ensembl Gene: ENSG00000135517 | OMIM ID: 154050 |
| Drug and gene relationship at DGIdb | |||
Expression of MIP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MIP | 4284 | 220863_at | -0.0701 | 0.7661 | |
| GSE20347 | MIP | 4284 | 220863_at | 0.0001 | 0.9989 | |
| GSE23400 | MIP | 4284 | 220863_at | -0.0677 | 0.0076 | |
| GSE26886 | MIP | 4284 | 220863_at | -0.0065 | 0.9670 | |
| GSE29001 | MIP | 4284 | 220863_at | -0.1899 | 0.1867 | |
| GSE38129 | MIP | 4284 | 220863_at | -0.0639 | 0.2118 | |
| GSE45670 | MIP | 4284 | 220863_at | -0.0170 | 0.8755 | |
| GSE53622 | MIP | 4284 | 24390 | 0.0937 | 0.4288 | |
| GSE53624 | MIP | 4284 | 24390 | -0.0130 | 0.9005 | |
| GSE63941 | MIP | 4284 | 220863_at | 0.0571 | 0.7181 | |
| GSE77861 | MIP | 4284 | 220863_at | -0.1883 | 0.1104 | |
| TCGA | MIP | 4284 | RNAseq | 2.4863 | 0.0075 |
Upregulated datasets: 1; Downregulated datasets: 0.
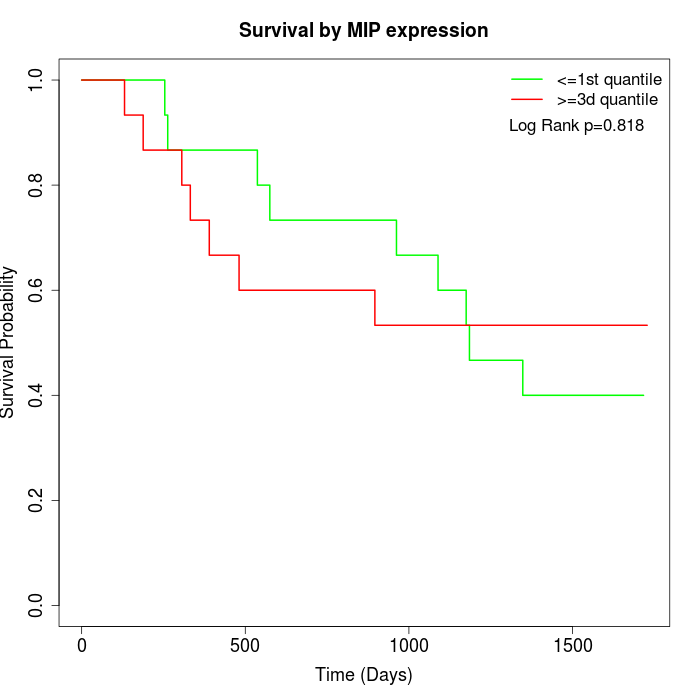
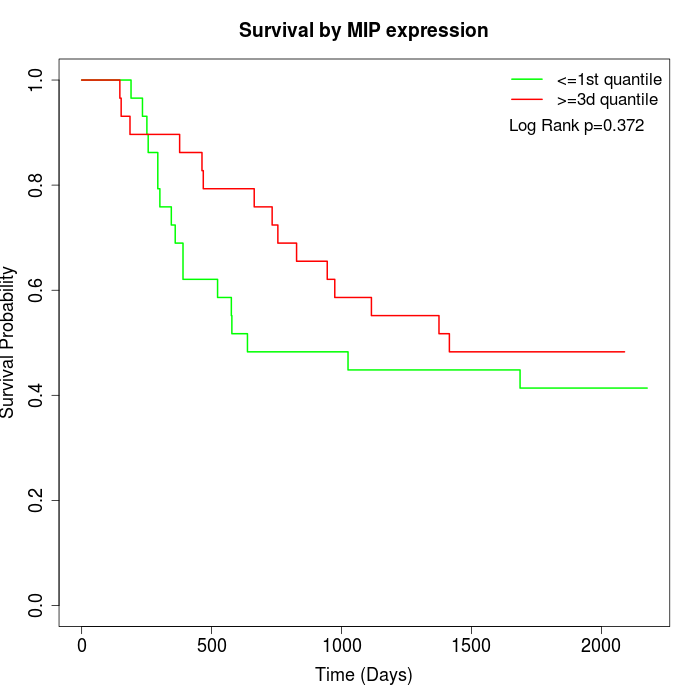
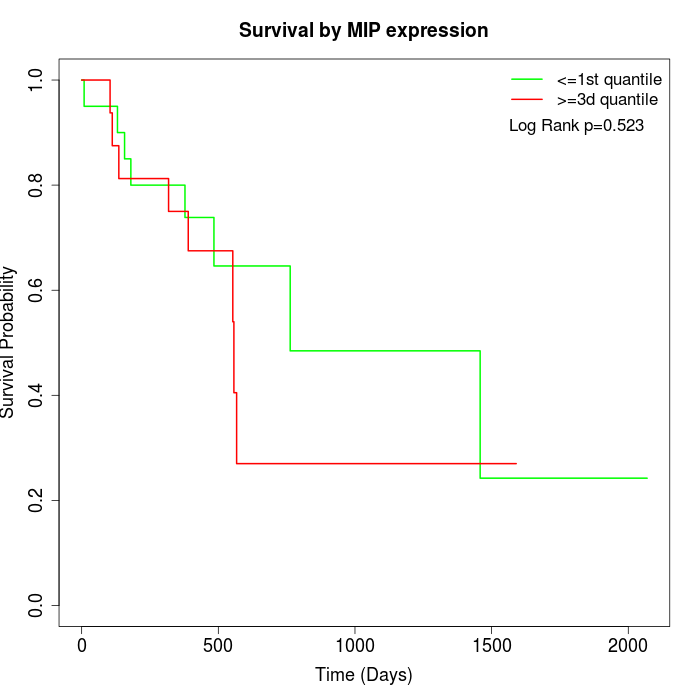
Survival by MIP expression:
Note: Click image to view full size file.
Copy number change of MIP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MIP | 4284 | 7 | 2 | 21 | |
| GSE20123 | MIP | 4284 | 7 | 1 | 22 | |
| GSE43470 | MIP | 4284 | 3 | 0 | 40 | |
| GSE46452 | MIP | 4284 | 8 | 1 | 50 | |
| GSE47630 | MIP | 4284 | 10 | 2 | 28 | |
| GSE54993 | MIP | 4284 | 0 | 5 | 65 | |
| GSE54994 | MIP | 4284 | 4 | 1 | 48 | |
| GSE60625 | MIP | 4284 | 0 | 0 | 11 | |
| GSE74703 | MIP | 4284 | 3 | 0 | 33 | |
| GSE74704 | MIP | 4284 | 4 | 1 | 15 | |
| TCGA | MIP | 4284 | 14 | 10 | 72 |
Total number of gains: 60; Total number of losses: 23; Total Number of normals: 405.
Somatic mutations of MIP:
Generating mutation plots.
Highly correlated genes for MIP:
Showing top 20/166 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MIP | TEKT2 | 0.68908 | 3 | 0 | 3 |
| MIP | ST8SIA2 | 0.675788 | 4 | 0 | 3 |
| MIP | C1orf61 | 0.671662 | 3 | 0 | 3 |
| MIP | ITIH1 | 0.661794 | 3 | 0 | 3 |
| MIP | PSG4 | 0.657309 | 3 | 0 | 3 |
| MIP | AMACR | 0.648331 | 4 | 0 | 3 |
| MIP | HBM | 0.634118 | 3 | 0 | 3 |
| MIP | OR3A2 | 0.631147 | 3 | 0 | 3 |
| MIP | ENHO | 0.629907 | 4 | 0 | 3 |
| MIP | HCG9 | 0.628727 | 4 | 0 | 3 |
| MIP | AGRP | 0.627398 | 3 | 0 | 3 |
| MIP | ZGLP1 | 0.626705 | 3 | 0 | 3 |
| MIP | HCN1 | 0.625846 | 4 | 0 | 3 |
| MIP | PRM2 | 0.624395 | 4 | 0 | 4 |
| MIP | STH | 0.616457 | 3 | 0 | 3 |
| MIP | KCNK17 | 0.613812 | 3 | 0 | 3 |
| MIP | PDYN | 0.613277 | 4 | 0 | 3 |
| MIP | RORC | 0.612241 | 4 | 0 | 3 |
| MIP | DNAAF1 | 0.610558 | 5 | 0 | 4 |
| MIP | OCM2 | 0.604733 | 4 | 0 | 4 |
For details and further investigation, click here