| Full name: hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 | Alias Symbol: BCNG-2|HAC-1 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 610 | HGNC ID: HGNC:4846 | Ensembl Gene: ENSG00000099822 | OMIM ID: 602781 |
| Drug and gene relationship at DGIdb | |||
HCN2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway |
Expression of HCN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCN2 | 610 | 214893_x_at | -0.1299 | 0.6104 | |
| GSE20347 | HCN2 | 610 | 214893_x_at | -0.1043 | 0.1459 | |
| GSE23400 | HCN2 | 610 | 214893_x_at | -0.1145 | 0.0000 | |
| GSE26886 | HCN2 | 610 | 214893_x_at | 0.2130 | 0.0136 | |
| GSE29001 | HCN2 | 610 | 214893_x_at | -0.1024 | 0.3322 | |
| GSE38129 | HCN2 | 610 | 214893_x_at | -0.1056 | 0.0667 | |
| GSE45670 | HCN2 | 610 | 214893_x_at | 0.0041 | 0.9645 | |
| GSE53622 | HCN2 | 610 | 53657 | -0.1726 | 0.0015 | |
| GSE53624 | HCN2 | 610 | 53657 | 0.0819 | 0.1529 | |
| GSE63941 | HCN2 | 610 | 214893_x_at | 0.0614 | 0.6877 | |
| GSE77861 | HCN2 | 610 | 214893_x_at | -0.1845 | 0.3464 | |
| GSE97050 | HCN2 | 610 | A_33_P3279640 | -0.0103 | 0.9733 | |
| SRP007169 | HCN2 | 610 | RNAseq | -6.0588 | 0.0000 | |
| SRP008496 | HCN2 | 610 | RNAseq | -4.5851 | 0.0000 | |
| SRP064894 | HCN2 | 610 | RNAseq | -2.6994 | 0.0000 | |
| SRP133303 | HCN2 | 610 | RNAseq | -1.7966 | 0.0000 | |
| SRP159526 | HCN2 | 610 | RNAseq | -2.9231 | 0.0000 | |
| SRP219564 | HCN2 | 610 | RNAseq | -1.6238 | 0.0241 | |
| TCGA | HCN2 | 610 | RNAseq | -0.2390 | 0.3398 |
Upregulated datasets: 0; Downregulated datasets: 6.
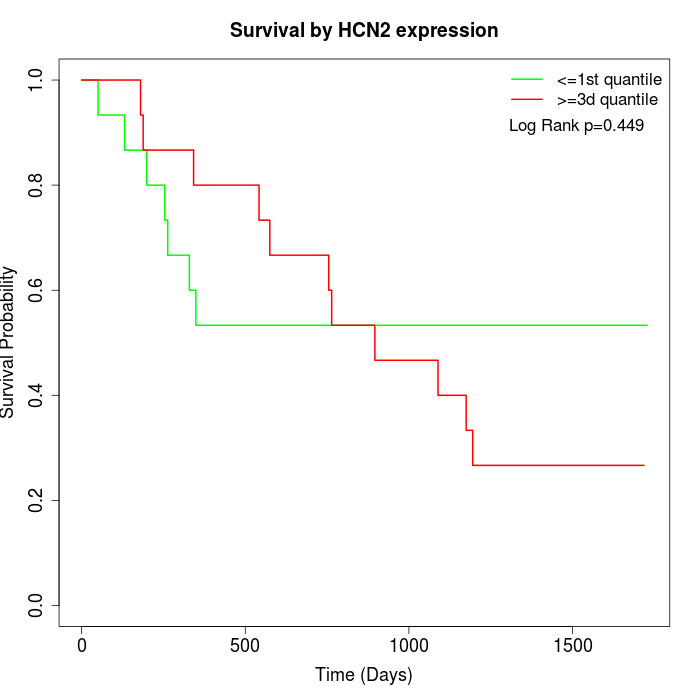
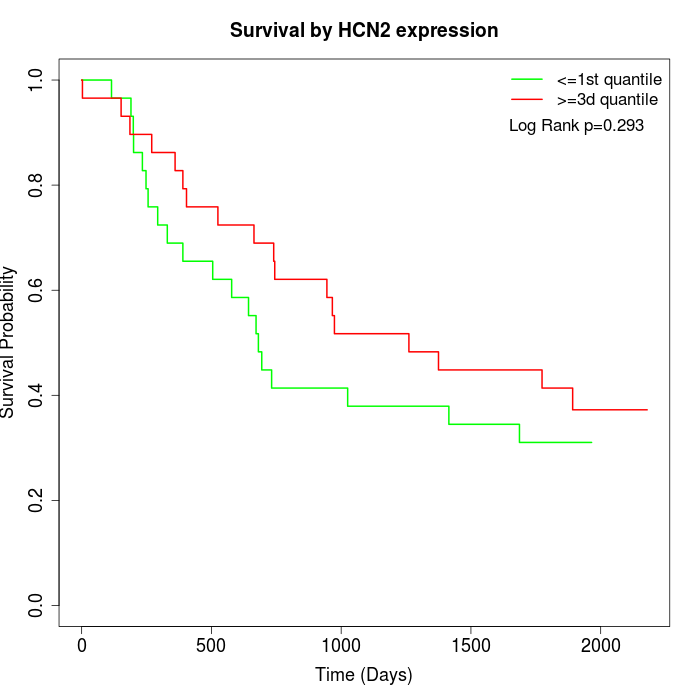
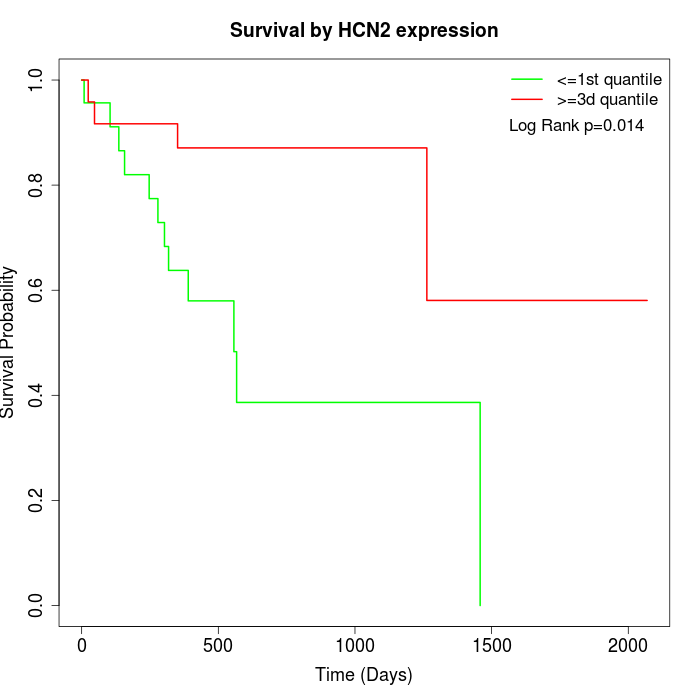
Survival by HCN2 expression:
Note: Click image to view full size file.
Copy number change of HCN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCN2 | 610 | 4 | 6 | 20 | |
| GSE20123 | HCN2 | 610 | 4 | 5 | 21 | |
| GSE43470 | HCN2 | 610 | 1 | 10 | 32 | |
| GSE46452 | HCN2 | 610 | 47 | 1 | 11 | |
| GSE47630 | HCN2 | 610 | 5 | 7 | 28 | |
| GSE54993 | HCN2 | 610 | 13 | 4 | 53 | |
| GSE54994 | HCN2 | 610 | 8 | 15 | 30 | |
| GSE60625 | HCN2 | 610 | 9 | 0 | 2 | |
| GSE74703 | HCN2 | 610 | 1 | 7 | 28 | |
| GSE74704 | HCN2 | 610 | 3 | 3 | 14 | |
| TCGA | HCN2 | 610 | 8 | 25 | 63 |
Total number of gains: 103; Total number of losses: 83; Total Number of normals: 302.
Somatic mutations of HCN2:
Generating mutation plots.
Highly correlated genes for HCN2:
Showing top 20/152 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCN2 | CRB3 | 0.683829 | 3 | 0 | 3 |
| HCN2 | ARHGAP30 | 0.671616 | 3 | 0 | 3 |
| HCN2 | ISLR2 | 0.669322 | 3 | 0 | 3 |
| HCN2 | LRRC4B | 0.664111 | 3 | 0 | 3 |
| HCN2 | TTLL9 | 0.658287 | 3 | 0 | 3 |
| HCN2 | MATN4 | 0.656117 | 3 | 0 | 3 |
| HCN2 | C16orf86 | 0.6546 | 3 | 0 | 3 |
| HCN2 | MEGF11 | 0.64375 | 3 | 0 | 3 |
| HCN2 | DUOXA2 | 0.643142 | 3 | 0 | 3 |
| HCN2 | NEU4 | 0.641754 | 3 | 0 | 3 |
| HCN2 | NKAIN4 | 0.63847 | 3 | 0 | 3 |
| HCN2 | CABP7 | 0.628874 | 3 | 0 | 3 |
| HCN2 | ZP1 | 0.627742 | 4 | 0 | 3 |
| HCN2 | JSRP1 | 0.619687 | 5 | 0 | 3 |
| HCN2 | AMBRA1 | 0.615441 | 4 | 0 | 3 |
| HCN2 | CHRNE | 0.614617 | 4 | 0 | 3 |
| HCN2 | DRD4 | 0.613768 | 6 | 0 | 5 |
| HCN2 | BRF1 | 0.61365 | 4 | 0 | 3 |
| HCN2 | ZNF646 | 0.612727 | 4 | 0 | 4 |
| HCN2 | OR1J4 | 0.612721 | 4 | 0 | 3 |
For details and further investigation, click here