| Full name: canopy FGF signaling regulator 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q36.3 | ||
| Entrez ID: 285888 | HGNC ID: HGNC:27786 | Ensembl Gene: ENSG00000146910 | OMIM ID: 612493 |
| Drug and gene relationship at DGIdb | |||
Expression of CNPY1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CNPY1 | 285888 | 1559283_a_at | -0.1040 | 0.6051 | |
| GSE26886 | CNPY1 | 285888 | 1559283_a_at | 0.0831 | 0.3550 | |
| GSE45670 | CNPY1 | 285888 | 1559283_a_at | -0.0065 | 0.9367 | |
| GSE53622 | CNPY1 | 285888 | 118021 | 0.0247 | 0.8804 | |
| GSE53624 | CNPY1 | 285888 | 118021 | -0.0650 | 0.5034 | |
| GSE63941 | CNPY1 | 285888 | 1559283_a_at | 0.0447 | 0.7731 | |
| GSE77861 | CNPY1 | 285888 | 1559283_a_at | -0.0441 | 0.6262 | |
| GSE97050 | CNPY1 | 285888 | A_33_P3220347 | 0.2423 | 0.3710 | |
| TCGA | CNPY1 | 285888 | RNAseq | 1.6711 | 0.1955 |
Upregulated datasets: 0; Downregulated datasets: 0.
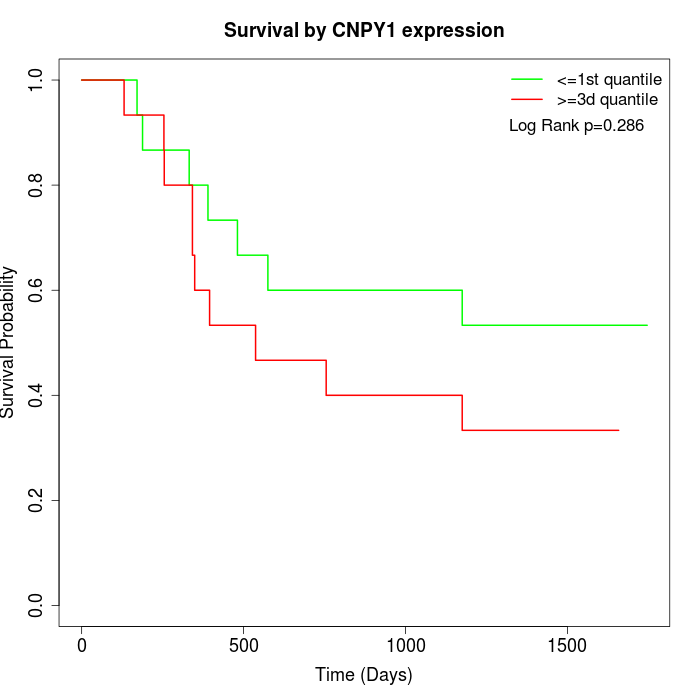
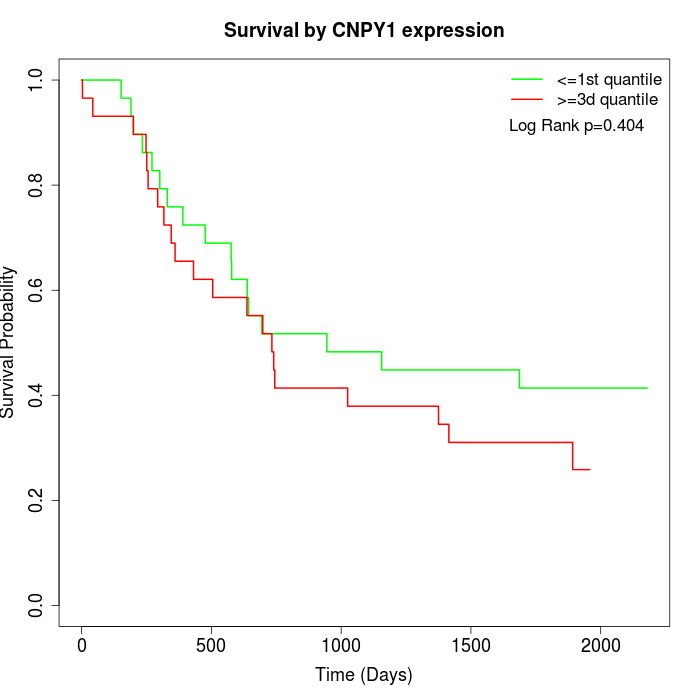
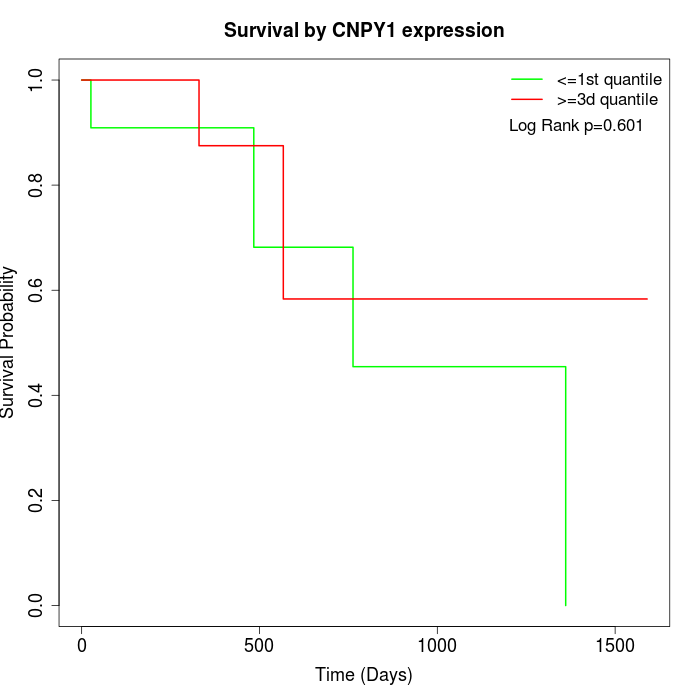
Survival by CNPY1 expression:
Note: Click image to view full size file.
Copy number change of CNPY1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CNPY1 | 285888 | 3 | 3 | 24 | |
| GSE20123 | CNPY1 | 285888 | 2 | 3 | 25 | |
| GSE43470 | CNPY1 | 285888 | 2 | 5 | 36 | |
| GSE46452 | CNPY1 | 285888 | 7 | 2 | 50 | |
| GSE47630 | CNPY1 | 285888 | 6 | 9 | 25 | |
| GSE54993 | CNPY1 | 285888 | 3 | 3 | 64 | |
| GSE54994 | CNPY1 | 285888 | 6 | 8 | 39 | |
| GSE60625 | CNPY1 | 285888 | 0 | 0 | 11 | |
| GSE74703 | CNPY1 | 285888 | 2 | 4 | 30 | |
| GSE74704 | CNPY1 | 285888 | 0 | 3 | 17 | |
| TCGA | CNPY1 | 285888 | 27 | 26 | 43 |
Total number of gains: 58; Total number of losses: 66; Total Number of normals: 364.
Somatic mutations of CNPY1:
Generating mutation plots.
Highly correlated genes for CNPY1:
Showing top 20/173 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CNPY1 | HCRT | 0.735638 | 4 | 0 | 3 |
| CNPY1 | FUZ | 0.731408 | 3 | 0 | 3 |
| CNPY1 | SSTR2 | 0.727365 | 4 | 0 | 4 |
| CNPY1 | FGF18 | 0.715391 | 3 | 0 | 3 |
| CNPY1 | LCN8 | 0.712571 | 3 | 0 | 3 |
| CNPY1 | PRSS38 | 0.710995 | 3 | 0 | 3 |
| CNPY1 | YY2 | 0.708158 | 3 | 0 | 3 |
| CNPY1 | DEFB123 | 0.702132 | 5 | 0 | 4 |
| CNPY1 | KCNQ4 | 0.70199 | 4 | 0 | 4 |
| CNPY1 | PRSS54 | 0.697595 | 3 | 0 | 3 |
| CNPY1 | DRGX | 0.694925 | 3 | 0 | 3 |
| CNPY1 | DCD | 0.69138 | 3 | 0 | 3 |
| CNPY1 | STRC | 0.691222 | 3 | 0 | 3 |
| CNPY1 | BCL2L10 | 0.691113 | 3 | 0 | 3 |
| CNPY1 | TNFRSF14 | 0.69002 | 3 | 0 | 3 |
| CNPY1 | LDHAL6A | 0.686154 | 3 | 0 | 3 |
| CNPY1 | JAKMIP1 | 0.685427 | 3 | 0 | 3 |
| CNPY1 | MAT1A | 0.67598 | 3 | 0 | 3 |
| CNPY1 | OR2C1 | 0.675173 | 5 | 0 | 4 |
| CNPY1 | PRAMEF5 | 0.675097 | 4 | 0 | 3 |
For details and further investigation, click here