| Full name: inhibin subunit beta C | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.3 | ||
| Entrez ID: 3626 | HGNC ID: HGNC:6068 | Ensembl Gene: ENSG00000175189 | OMIM ID: 601233 |
| Drug and gene relationship at DGIdb | |||
INHBC involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells |
Expression of INHBC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE23400 | INHBC | 3626 | 207687_at | -0.2107 | 0.0000 | |
| GSE53622 | INHBC | 3626 | 19803 | 0.4548 | 0.0000 | |
| GSE53624 | INHBC | 3626 | 19803 | 0.4489 | 0.0000 | |
| GSE77861 | INHBC | 3626 | 207687_at | -0.1873 | 0.2035 | |
| GSE97050 | INHBC | 3626 | A_24_P155502 | -0.0762 | 0.7968 | |
| TCGA | INHBC | 3626 | RNAseq | 3.3040 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
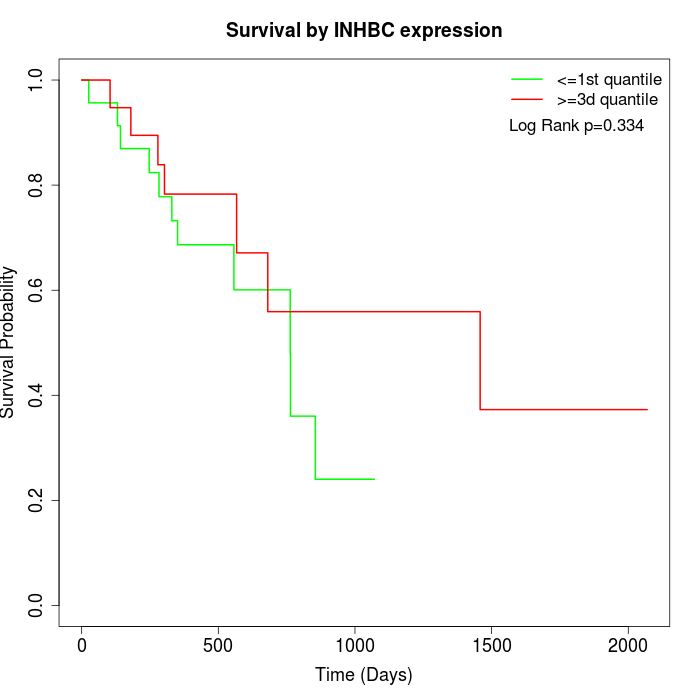
Survival by INHBC expression:
Note: Click image to view full size file.
Copy number change of INHBC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INHBC | 3626 | 6 | 1 | 23 | |
| GSE20123 | INHBC | 3626 | 6 | 0 | 24 | |
| GSE43470 | INHBC | 3626 | 2 | 0 | 41 | |
| GSE46452 | INHBC | 3626 | 8 | 1 | 50 | |
| GSE47630 | INHBC | 3626 | 10 | 2 | 28 | |
| GSE54993 | INHBC | 3626 | 0 | 5 | 65 | |
| GSE54994 | INHBC | 3626 | 4 | 1 | 48 | |
| GSE60625 | INHBC | 3626 | 0 | 0 | 11 | |
| GSE74703 | INHBC | 3626 | 2 | 0 | 34 | |
| GSE74704 | INHBC | 3626 | 4 | 0 | 16 | |
| TCGA | INHBC | 3626 | 14 | 10 | 72 |
Total number of gains: 56; Total number of losses: 20; Total Number of normals: 412.
Somatic mutations of INHBC:
Generating mutation plots.
Highly correlated genes for INHBC:
Showing top 20/104 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INHBC | GUCY2C | 0.760172 | 3 | 0 | 3 |
| INHBC | AMHR2 | 0.752652 | 3 | 0 | 3 |
| INHBC | SYT2 | 0.74135 | 3 | 0 | 3 |
| INHBC | HNF1B | 0.727713 | 3 | 0 | 3 |
| INHBC | BEAN1 | 0.723658 | 3 | 0 | 3 |
| INHBC | GFRA2 | 0.72195 | 3 | 0 | 3 |
| INHBC | SSTR2 | 0.720379 | 3 | 0 | 3 |
| INHBC | HCRT | 0.716267 | 3 | 0 | 3 |
| INHBC | SLC22A14 | 0.711962 | 3 | 0 | 3 |
| INHBC | FASLG | 0.708851 | 3 | 0 | 3 |
| INHBC | DEFB108B | 0.70725 | 3 | 0 | 3 |
| INHBC | ITGA10 | 0.707239 | 3 | 0 | 3 |
| INHBC | OTOF | 0.703618 | 3 | 0 | 3 |
| INHBC | SV2C | 0.702131 | 3 | 0 | 3 |
| INHBC | MC5R | 0.700172 | 3 | 0 | 3 |
| INHBC | C3orf36 | 0.698204 | 3 | 0 | 3 |
| INHBC | MAP3K10 | 0.691526 | 3 | 0 | 3 |
| INHBC | KCNMB2 | 0.690158 | 3 | 0 | 3 |
| INHBC | APOM | 0.687427 | 3 | 0 | 3 |
| INHBC | NPY2R | 0.683409 | 3 | 0 | 3 |
For details and further investigation, click here