| Full name: heat shock protein 90 beta family member 1 | Alias Symbol: GP96|GRP94 | ||
| Type: protein-coding gene | Cytoband: 12q23.3 | ||
| Entrez ID: 7184 | HGNC ID: HGNC:12028 | Ensembl Gene: ENSG00000166598 | OMIM ID: 191175 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
HSP90B1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04621 | NOD-like receptor signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05215 | Prostate cancer |
Expression of HSP90B1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HSP90B1 | 7184 | 216449_x_at | 0.5453 | 0.3227 | |
| GSE20347 | HSP90B1 | 7184 | 216449_x_at | 0.3150 | 0.0101 | |
| GSE23400 | HSP90B1 | 7184 | 216450_x_at | 0.2863 | 0.0009 | |
| GSE26886 | HSP90B1 | 7184 | 216449_x_at | 0.5872 | 0.0208 | |
| GSE29001 | HSP90B1 | 7184 | 216449_x_at | 0.7049 | 0.1183 | |
| GSE38129 | HSP90B1 | 7184 | 216449_x_at | 0.6156 | 0.0010 | |
| GSE45670 | HSP90B1 | 7184 | 216449_x_at | 0.7805 | 0.0001 | |
| GSE53622 | HSP90B1 | 7184 | 96817 | 0.9596 | 0.0000 | |
| GSE53624 | HSP90B1 | 7184 | 96817 | 1.2497 | 0.0000 | |
| GSE63941 | HSP90B1 | 7184 | 216449_x_at | -0.2739 | 0.4433 | |
| GSE77861 | HSP90B1 | 7184 | 216449_x_at | 0.7585 | 0.0096 | |
| GSE97050 | HSP90B1 | 7184 | A_23_P2601 | 0.7727 | 0.0750 | |
| SRP007169 | HSP90B1 | 7184 | RNAseq | 1.1688 | 0.0013 | |
| SRP008496 | HSP90B1 | 7184 | RNAseq | 1.3531 | 0.0000 | |
| SRP064894 | HSP90B1 | 7184 | RNAseq | 0.9864 | 0.0000 | |
| SRP133303 | HSP90B1 | 7184 | RNAseq | 1.2056 | 0.0000 | |
| SRP159526 | HSP90B1 | 7184 | RNAseq | 1.3144 | 0.0000 | |
| SRP193095 | HSP90B1 | 7184 | RNAseq | 0.4799 | 0.0018 | |
| SRP219564 | HSP90B1 | 7184 | RNAseq | 1.0483 | 0.0088 | |
| TCGA | HSP90B1 | 7184 | RNAseq | 0.0817 | 0.0249 |
Upregulated datasets: 6; Downregulated datasets: 0.
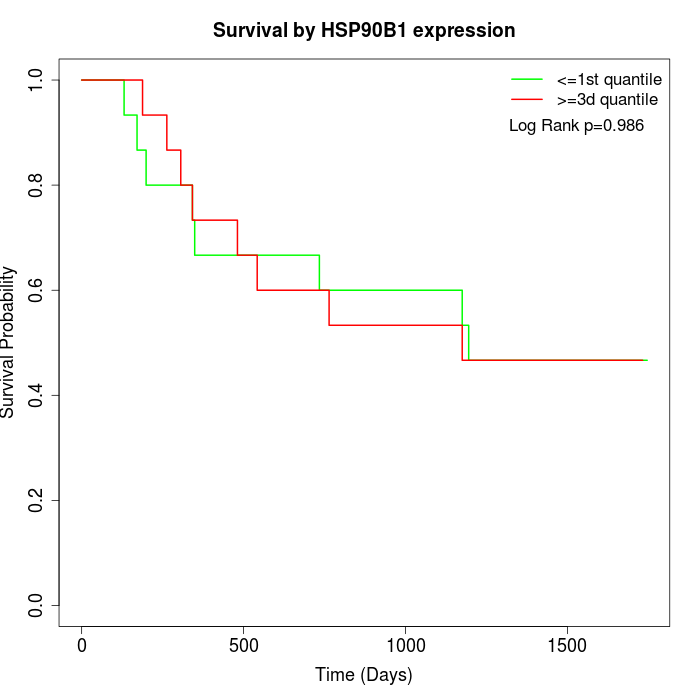
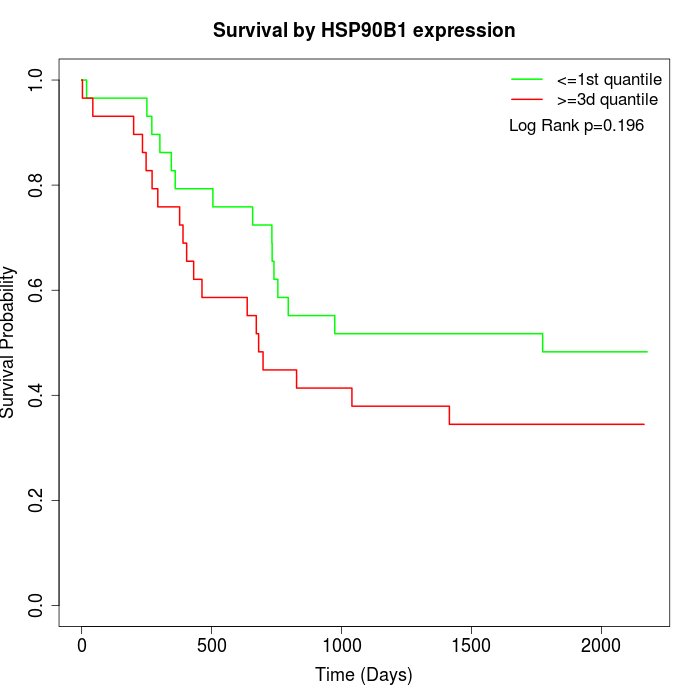
Survival by HSP90B1 expression:
Note: Click image to view full size file.
Copy number change of HSP90B1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HSP90B1 | 7184 | 4 | 2 | 24 | |
| GSE20123 | HSP90B1 | 7184 | 4 | 2 | 24 | |
| GSE43470 | HSP90B1 | 7184 | 2 | 0 | 41 | |
| GSE46452 | HSP90B1 | 7184 | 9 | 1 | 49 | |
| GSE47630 | HSP90B1 | 7184 | 9 | 1 | 30 | |
| GSE54993 | HSP90B1 | 7184 | 0 | 5 | 65 | |
| GSE54994 | HSP90B1 | 7184 | 4 | 2 | 47 | |
| GSE60625 | HSP90B1 | 7184 | 0 | 0 | 11 | |
| GSE74703 | HSP90B1 | 7184 | 2 | 0 | 34 | |
| GSE74704 | HSP90B1 | 7184 | 1 | 2 | 17 | |
| TCGA | HSP90B1 | 7184 | 19 | 11 | 66 |
Total number of gains: 54; Total number of losses: 26; Total Number of normals: 408.
Somatic mutations of HSP90B1:
Generating mutation plots.
Highly correlated genes for HSP90B1:
Showing top 20/1534 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HSP90B1 | PYCR2 | 0.785575 | 3 | 0 | 3 |
| HSP90B1 | SLC19A1 | 0.750925 | 3 | 0 | 3 |
| HSP90B1 | CLPTM1L | 0.731844 | 6 | 0 | 6 |
| HSP90B1 | SPON2 | 0.731443 | 3 | 0 | 3 |
| HSP90B1 | GBP5 | 0.7246 | 3 | 0 | 3 |
| HSP90B1 | FAM89A | 0.721075 | 3 | 0 | 3 |
| HSP90B1 | NOP56 | 0.716888 | 3 | 0 | 3 |
| HSP90B1 | ZIC5 | 0.715869 | 3 | 0 | 3 |
| HSP90B1 | PDIA5 | 0.71274 | 4 | 0 | 3 |
| HSP90B1 | BANF1 | 0.708105 | 5 | 0 | 5 |
| HSP90B1 | GRINA | 0.702207 | 5 | 0 | 5 |
| HSP90B1 | CCDC127 | 0.702197 | 3 | 0 | 3 |
| HSP90B1 | CMSS1 | 0.69749 | 6 | 0 | 6 |
| HSP90B1 | PPIB | 0.696536 | 7 | 0 | 6 |
| HSP90B1 | FAM91A1 | 0.696238 | 6 | 0 | 6 |
| HSP90B1 | BSDC1 | 0.695637 | 3 | 0 | 3 |
| HSP90B1 | EBNA1BP2 | 0.693578 | 3 | 0 | 3 |
| HSP90B1 | DCBLD1 | 0.69279 | 6 | 0 | 6 |
| HSP90B1 | RAB34 | 0.69212 | 3 | 0 | 3 |
| HSP90B1 | ELOF1 | 0.688969 | 3 | 0 | 3 |
For details and further investigation, click here