| Full name: pyrroline-5-carboxylate reductase 2 | Alias Symbol: P5CR2 | ||
| Type: protein-coding gene | Cytoband: 1q42.12 | ||
| Entrez ID: 29920 | HGNC ID: HGNC:30262 | Ensembl Gene: ENSG00000143811 | OMIM ID: 616406 |
| Drug and gene relationship at DGIdb | |||
Expression of PYCR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | PYCR2 | 29920 | 4893 | 0.9700 | 0.0000 | |
| GSE53624 | PYCR2 | 29920 | 4893 | 1.0130 | 0.0000 | |
| GSE97050 | PYCR2 | 29920 | A_23_P149042 | 0.5824 | 0.1189 | |
| SRP007169 | PYCR2 | 29920 | RNAseq | 1.6605 | 0.0000 | |
| SRP008496 | PYCR2 | 29920 | RNAseq | 1.5380 | 0.0000 | |
| SRP064894 | PYCR2 | 29920 | RNAseq | 1.0493 | 0.0000 | |
| SRP133303 | PYCR2 | 29920 | RNAseq | 1.0081 | 0.0000 | |
| SRP159526 | PYCR2 | 29920 | RNAseq | 1.0903 | 0.0094 | |
| SRP193095 | PYCR2 | 29920 | RNAseq | 1.2405 | 0.0000 | |
| SRP219564 | PYCR2 | 29920 | RNAseq | 0.8365 | 0.0141 | |
| TCGA | PYCR2 | 29920 | RNAseq | 0.2494 | 0.0001 |
Upregulated datasets: 7; Downregulated datasets: 0.
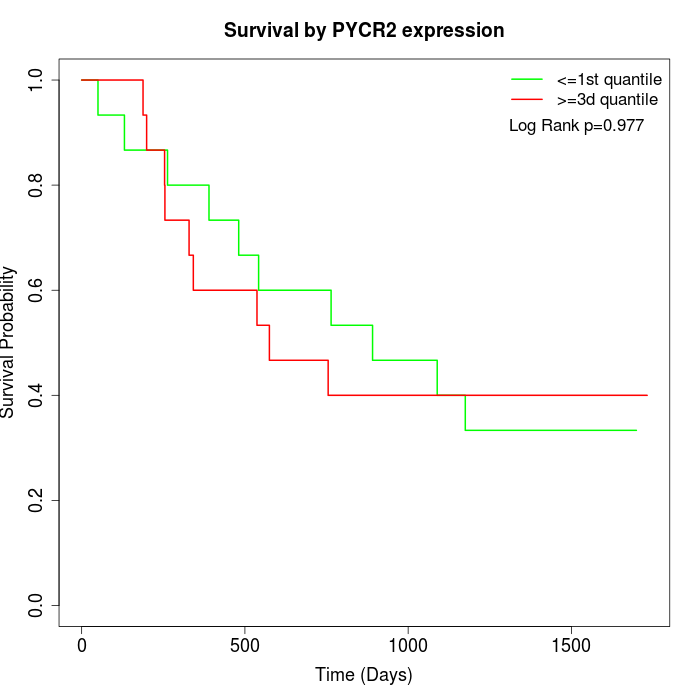
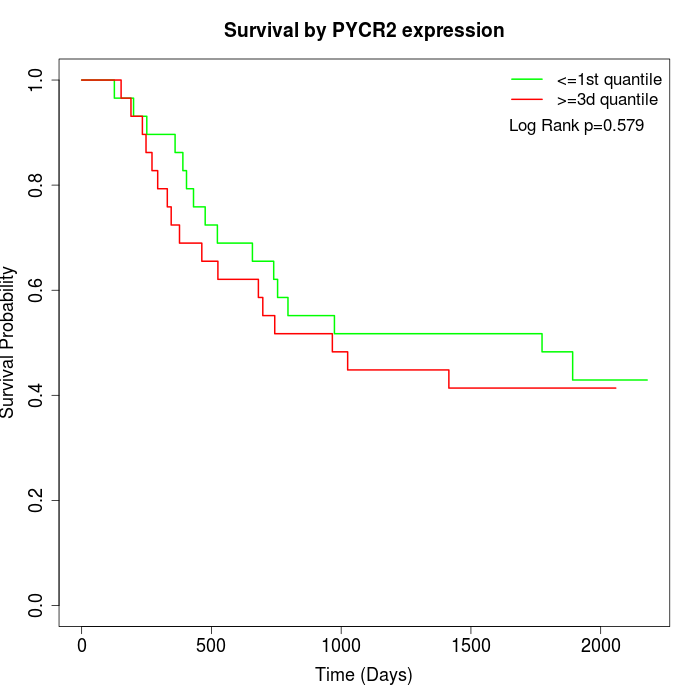
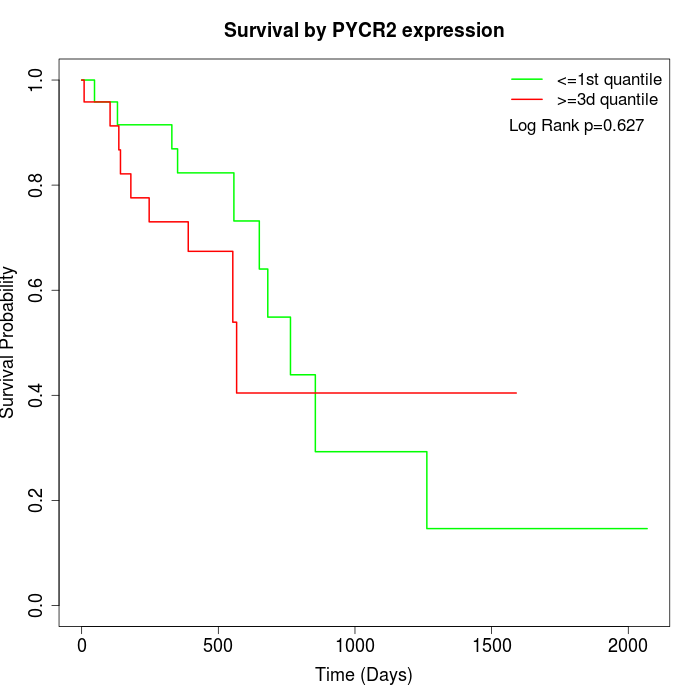
Survival by PYCR2 expression:
Note: Click image to view full size file.
Copy number change of PYCR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYCR2 | 29920 | 11 | 0 | 19 | |
| GSE20123 | PYCR2 | 29920 | 11 | 0 | 19 | |
| GSE43470 | PYCR2 | 29920 | 7 | 1 | 35 | |
| GSE46452 | PYCR2 | 29920 | 3 | 1 | 55 | |
| GSE47630 | PYCR2 | 29920 | 15 | 0 | 25 | |
| GSE54993 | PYCR2 | 29920 | 0 | 6 | 64 | |
| GSE54994 | PYCR2 | 29920 | 16 | 0 | 37 | |
| GSE60625 | PYCR2 | 29920 | 0 | 0 | 11 | |
| GSE74703 | PYCR2 | 29920 | 7 | 1 | 28 | |
| GSE74704 | PYCR2 | 29920 | 5 | 0 | 15 | |
| TCGA | PYCR2 | 29920 | 46 | 3 | 47 |
Total number of gains: 121; Total number of losses: 12; Total Number of normals: 355.
Somatic mutations of PYCR2:
Generating mutation plots.
Highly correlated genes for PYCR2:
Showing top 20/925 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYCR2 | XPR1 | 0.837511 | 3 | 0 | 3 |
| PYCR2 | ZNF281 | 0.830769 | 3 | 0 | 3 |
| PYCR2 | RAI14 | 0.815232 | 3 | 0 | 3 |
| PYCR2 | XPO1 | 0.813164 | 3 | 0 | 3 |
| PYCR2 | BOLA3 | 0.812372 | 3 | 0 | 3 |
| PYCR2 | ECE1 | 0.809583 | 3 | 0 | 3 |
| PYCR2 | ELK4 | 0.807369 | 3 | 0 | 3 |
| PYCR2 | BRIX1 | 0.805581 | 3 | 0 | 3 |
| PYCR2 | LIMK1 | 0.803903 | 3 | 0 | 3 |
| PYCR2 | SIX4 | 0.803454 | 3 | 0 | 3 |
| PYCR2 | STC2 | 0.802388 | 3 | 0 | 3 |
| PYCR2 | TMEM209 | 0.802135 | 3 | 0 | 3 |
| PYCR2 | MAD2L2 | 0.799816 | 3 | 0 | 3 |
| PYCR2 | PFDN2 | 0.799365 | 3 | 0 | 3 |
| PYCR2 | PDIA4 | 0.798052 | 3 | 0 | 3 |
| PYCR2 | WDR54 | 0.798004 | 3 | 0 | 3 |
| PYCR2 | MTHFD2 | 0.795842 | 3 | 0 | 3 |
| PYCR2 | MARCKSL1 | 0.795818 | 3 | 0 | 3 |
| PYCR2 | RPN1 | 0.795484 | 3 | 0 | 3 |
| PYCR2 | FUS | 0.79516 | 3 | 0 | 3 |
For details and further investigation, click here