| Full name: heat shock protein family A (Hsp70) member 9 | Alias Symbol: GRP75|PBP74|mot-2|mthsp75 | ||
| Type: protein-coding gene | Cytoband: 5q31.2 | ||
| Entrez ID: 3313 | HGNC ID: HGNC:5244 | Ensembl Gene: ENSG00000113013 | OMIM ID: 600548 |
| Drug and gene relationship at DGIdb | |||
HSPA9 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05152 | Tuberculosis |
Expression of HSPA9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HSPA9 | 3313 | 200691_s_at | 0.1159 | 0.8567 | |
| GSE20347 | HSPA9 | 3313 | 200691_s_at | 0.3729 | 0.0491 | |
| GSE23400 | HSPA9 | 3313 | 200691_s_at | 0.3946 | 0.0000 | |
| GSE26886 | HSPA9 | 3313 | 200691_s_at | -0.2275 | 0.1086 | |
| GSE29001 | HSPA9 | 3313 | 200691_s_at | -0.1664 | 0.6404 | |
| GSE38129 | HSPA9 | 3313 | 200691_s_at | 0.4307 | 0.0016 | |
| GSE45670 | HSPA9 | 3313 | 200691_s_at | 0.2758 | 0.1589 | |
| GSE53622 | HSPA9 | 3313 | 84724 | 0.1489 | 0.0081 | |
| GSE53624 | HSPA9 | 3313 | 84724 | 0.2857 | 0.0000 | |
| GSE63941 | HSPA9 | 3313 | 200691_s_at | 0.2227 | 0.6500 | |
| GSE77861 | HSPA9 | 3313 | 200691_s_at | 0.1519 | 0.5588 | |
| GSE97050 | HSPA9 | 3313 | A_24_P77676 | 0.1032 | 0.7514 | |
| SRP007169 | HSPA9 | 3313 | RNAseq | 0.5067 | 0.2258 | |
| SRP008496 | HSPA9 | 3313 | RNAseq | 0.2166 | 0.4260 | |
| SRP064894 | HSPA9 | 3313 | RNAseq | -0.0461 | 0.7899 | |
| SRP133303 | HSPA9 | 3313 | RNAseq | 0.8436 | 0.0014 | |
| SRP159526 | HSPA9 | 3313 | RNAseq | 0.4261 | 0.2570 | |
| SRP193095 | HSPA9 | 3313 | RNAseq | 0.0795 | 0.5177 | |
| SRP219564 | HSPA9 | 3313 | RNAseq | 0.2944 | 0.4464 | |
| TCGA | HSPA9 | 3313 | RNAseq | 0.0262 | 0.5204 |
Upregulated datasets: 0; Downregulated datasets: 0.
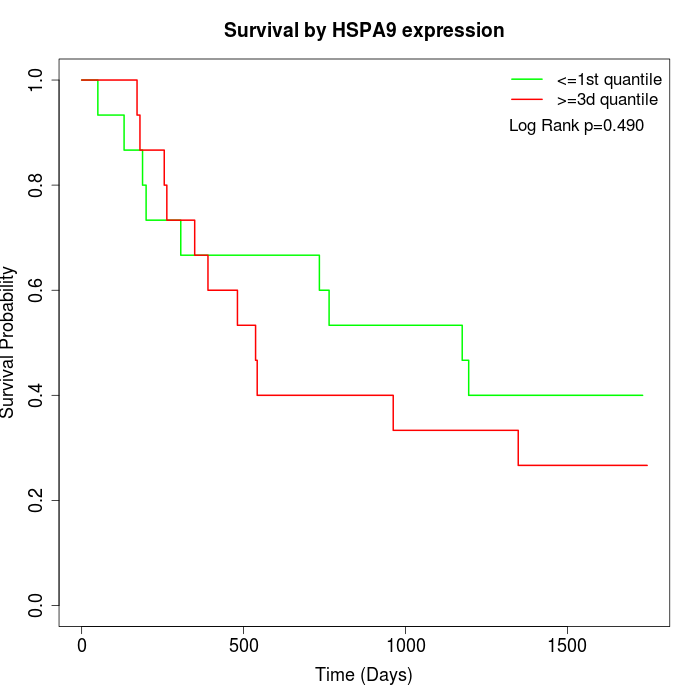
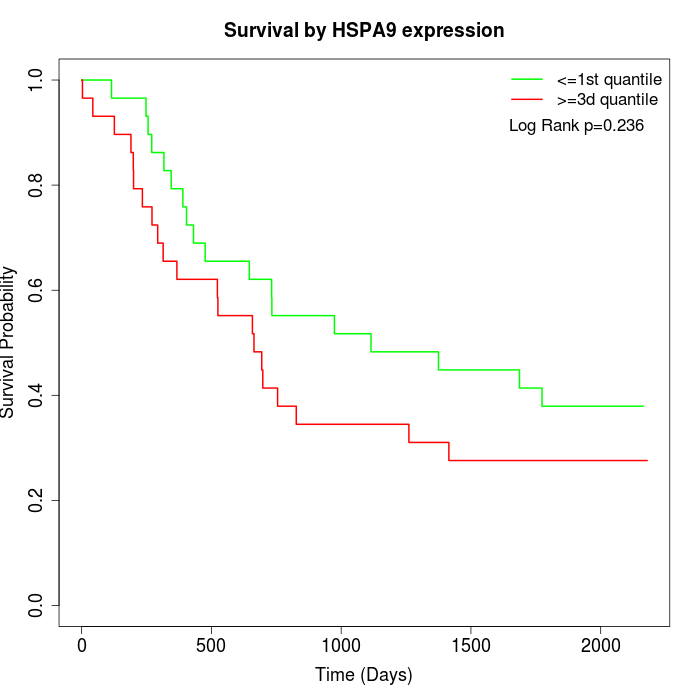
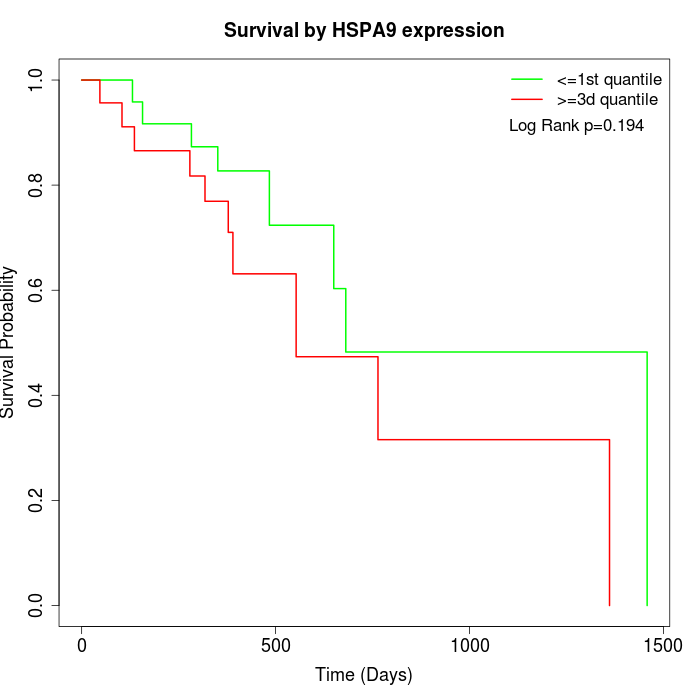
Survival by HSPA9 expression:
Note: Click image to view full size file.
Copy number change of HSPA9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HSPA9 | 3313 | 3 | 10 | 17 | |
| GSE20123 | HSPA9 | 3313 | 4 | 10 | 16 | |
| GSE43470 | HSPA9 | 3313 | 4 | 5 | 34 | |
| GSE46452 | HSPA9 | 3313 | 0 | 27 | 32 | |
| GSE47630 | HSPA9 | 3313 | 0 | 21 | 19 | |
| GSE54993 | HSPA9 | 3313 | 9 | 1 | 60 | |
| GSE54994 | HSPA9 | 3313 | 3 | 13 | 37 | |
| GSE60625 | HSPA9 | 3313 | 0 | 0 | 11 | |
| GSE74703 | HSPA9 | 3313 | 3 | 4 | 29 | |
| GSE74704 | HSPA9 | 3313 | 3 | 4 | 13 | |
| TCGA | HSPA9 | 3313 | 3 | 37 | 56 |
Total number of gains: 32; Total number of losses: 132; Total Number of normals: 324.
Somatic mutations of HSPA9:
Generating mutation plots.
Highly correlated genes for HSPA9:
Showing top 20/430 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HSPA9 | TMEM54 | 0.790635 | 3 | 0 | 3 |
| HSPA9 | ZBTB8OS | 0.789368 | 3 | 0 | 3 |
| HSPA9 | NFKB1 | 0.789314 | 3 | 0 | 3 |
| HSPA9 | NUDT9 | 0.78506 | 3 | 0 | 3 |
| HSPA9 | CETN3 | 0.771384 | 4 | 0 | 4 |
| HSPA9 | TMEM19 | 0.760485 | 3 | 0 | 3 |
| HSPA9 | FAHD1 | 0.759374 | 3 | 0 | 3 |
| HSPA9 | YIPF4 | 0.75907 | 4 | 0 | 4 |
| HSPA9 | ATG12 | 0.754271 | 3 | 0 | 3 |
| HSPA9 | UBLCP1 | 0.752928 | 4 | 0 | 4 |
| HSPA9 | C1orf226 | 0.751866 | 3 | 0 | 3 |
| HSPA9 | NBAS | 0.750373 | 3 | 0 | 3 |
| HSPA9 | ARFGEF2 | 0.740937 | 3 | 0 | 3 |
| HSPA9 | CASP4 | 0.738675 | 3 | 0 | 3 |
| HSPA9 | MMADHC | 0.733752 | 3 | 0 | 3 |
| HSPA9 | ING2 | 0.723999 | 3 | 0 | 3 |
| HSPA9 | AJUBA | 0.723248 | 3 | 0 | 3 |
| HSPA9 | RCHY1 | 0.722512 | 3 | 0 | 3 |
| HSPA9 | MRPL30 | 0.715183 | 4 | 0 | 3 |
| HSPA9 | KCMF1 | 0.706274 | 4 | 0 | 4 |
For details and further investigation, click here