| Full name: potassium channel modulatory factor 1 | Alias Symbol: DEBT91|PCMF|DKFZP434L1021|ZZZ1 | ||
| Type: protein-coding gene | Cytoband: 2p11.2 | ||
| Entrez ID: 56888 | HGNC ID: HGNC:20589 | Ensembl Gene: ENSG00000176407 | OMIM ID: 614719 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of KCMF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCMF1 | 56888 | 217938_s_at | -0.0688 | 0.8948 | |
| GSE20347 | KCMF1 | 56888 | 217938_s_at | -0.2309 | 0.0813 | |
| GSE23400 | KCMF1 | 56888 | 217938_s_at | -0.0589 | 0.4183 | |
| GSE26886 | KCMF1 | 56888 | 217938_s_at | -0.3433 | 0.0094 | |
| GSE29001 | KCMF1 | 56888 | 217938_s_at | -0.3010 | 0.0751 | |
| GSE38129 | KCMF1 | 56888 | 217938_s_at | -0.0282 | 0.8515 | |
| GSE45670 | KCMF1 | 56888 | 217938_s_at | -0.0721 | 0.6657 | |
| GSE53622 | KCMF1 | 56888 | 96164 | 0.0704 | 0.4367 | |
| GSE53624 | KCMF1 | 56888 | 96164 | 0.0806 | 0.3355 | |
| GSE63941 | KCMF1 | 56888 | 217938_s_at | 0.0413 | 0.9199 | |
| GSE77861 | KCMF1 | 56888 | 217938_s_at | -0.0661 | 0.7404 | |
| GSE97050 | KCMF1 | 56888 | A_33_P3354181 | -0.2349 | 0.4440 | |
| SRP007169 | KCMF1 | 56888 | RNAseq | -1.1581 | 0.0023 | |
| SRP008496 | KCMF1 | 56888 | RNAseq | -0.5519 | 0.0871 | |
| SRP064894 | KCMF1 | 56888 | RNAseq | -0.5615 | 0.0001 | |
| SRP133303 | KCMF1 | 56888 | RNAseq | 0.1646 | 0.2641 | |
| SRP159526 | KCMF1 | 56888 | RNAseq | 0.1932 | 0.3633 | |
| SRP193095 | KCMF1 | 56888 | RNAseq | -0.3010 | 0.0169 | |
| SRP219564 | KCMF1 | 56888 | RNAseq | -0.3182 | 0.4209 | |
| TCGA | KCMF1 | 56888 | RNAseq | 0.0874 | 0.0585 |
Upregulated datasets: 0; Downregulated datasets: 1.
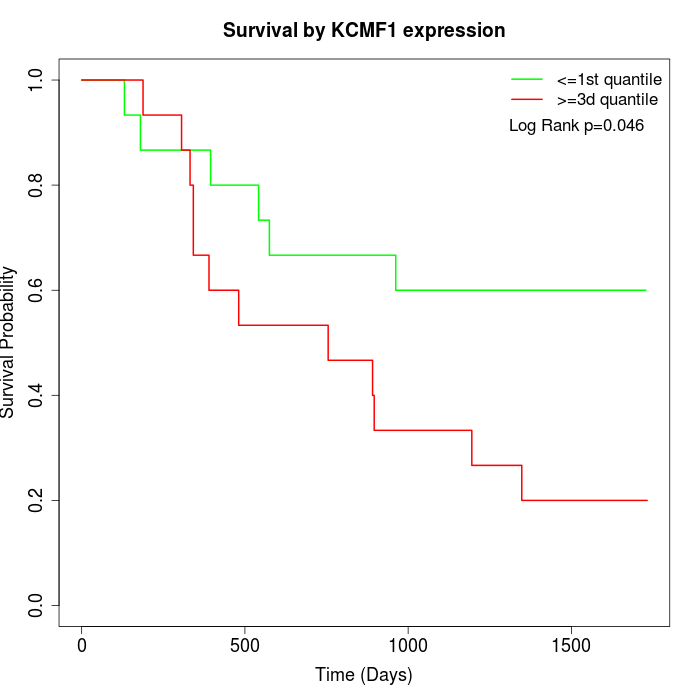
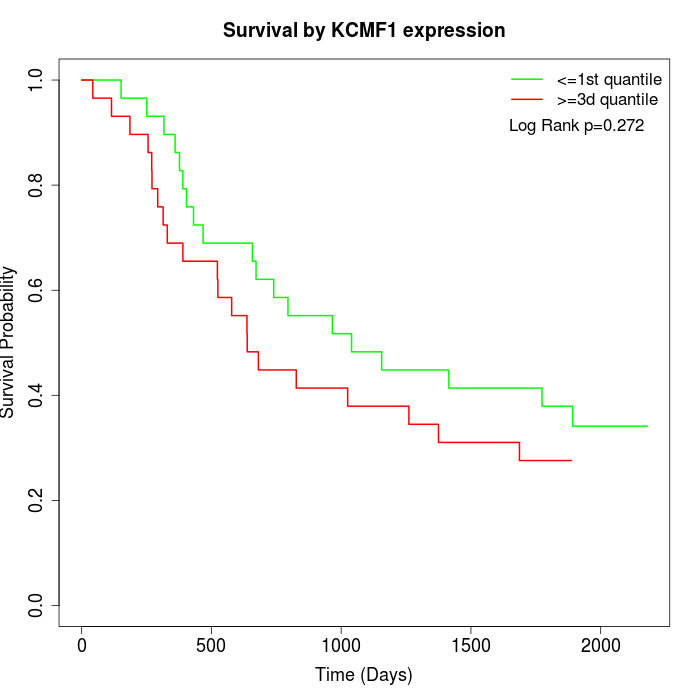
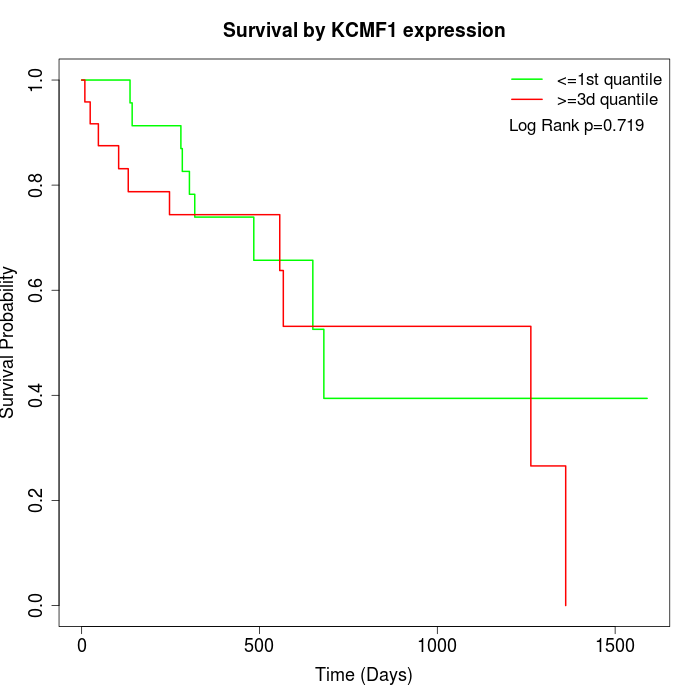
Survival by KCMF1 expression:
Note: Click image to view full size file.
Copy number change of KCMF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCMF1 | 56888 | 9 | 2 | 19 | |
| GSE20123 | KCMF1 | 56888 | 9 | 2 | 19 | |
| GSE43470 | KCMF1 | 56888 | 4 | 2 | 37 | |
| GSE46452 | KCMF1 | 56888 | 2 | 3 | 54 | |
| GSE47630 | KCMF1 | 56888 | 7 | 0 | 33 | |
| GSE54993 | KCMF1 | 56888 | 0 | 6 | 64 | |
| GSE54994 | KCMF1 | 56888 | 10 | 0 | 43 | |
| GSE60625 | KCMF1 | 56888 | 0 | 3 | 8 | |
| GSE74703 | KCMF1 | 56888 | 4 | 1 | 31 | |
| GSE74704 | KCMF1 | 56888 | 7 | 0 | 13 | |
| TCGA | KCMF1 | 56888 | 35 | 4 | 57 |
Total number of gains: 87; Total number of losses: 23; Total Number of normals: 378.
Somatic mutations of KCMF1:
Generating mutation plots.
Highly correlated genes for KCMF1:
Showing top 20/563 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCMF1 | SNX33 | 0.778275 | 3 | 0 | 3 |
| KCMF1 | ANKRD31 | 0.770917 | 3 | 0 | 3 |
| KCMF1 | UFSP2 | 0.767746 | 3 | 0 | 3 |
| KCMF1 | TMEM59 | 0.766353 | 3 | 0 | 3 |
| KCMF1 | EAF1 | 0.756198 | 3 | 0 | 3 |
| KCMF1 | SNRK | 0.744907 | 4 | 0 | 3 |
| KCMF1 | GANC | 0.74268 | 3 | 0 | 3 |
| KCMF1 | TBL1Y | 0.7402 | 3 | 0 | 3 |
| KCMF1 | LDHD | 0.726173 | 3 | 0 | 3 |
| KCMF1 | IPMK | 0.724183 | 4 | 0 | 4 |
| KCMF1 | STXBP5 | 0.715674 | 3 | 0 | 3 |
| KCMF1 | TTC23 | 0.714236 | 3 | 0 | 3 |
| KCMF1 | ZNF547 | 0.710643 | 3 | 0 | 3 |
| KCMF1 | THUMPD3 | 0.709475 | 4 | 0 | 3 |
| KCMF1 | LRRC28 | 0.708069 | 4 | 0 | 3 |
| KCMF1 | PSMD7 | 0.707606 | 4 | 0 | 3 |
| KCMF1 | HSPA9 | 0.706274 | 4 | 0 | 4 |
| KCMF1 | C9orf85 | 0.70611 | 4 | 0 | 3 |
| KCMF1 | KCTD9 | 0.705727 | 3 | 0 | 3 |
| KCMF1 | ESR2 | 0.703341 | 4 | 0 | 3 |
For details and further investigation, click here