| Full name: autophagy related 12 | Alias Symbol: APG12 | ||
| Type: protein-coding gene | Cytoband: 5q22.3 | ||
| Entrez ID: 9140 | HGNC ID: HGNC:588 | Ensembl Gene: ENSG00000145782 | OMIM ID: 609608 |
| Drug and gene relationship at DGIdb | |||
ATG12 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04622 | RIG-I-like receptor signaling pathway |
Expression of ATG12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATG12 | 9140 | 213026_at | -0.2596 | 0.6564 | |
| GSE20347 | ATG12 | 9140 | 213026_at | -0.3170 | 0.0377 | |
| GSE23400 | ATG12 | 9140 | 213026_at | 0.0991 | 0.5157 | |
| GSE26886 | ATG12 | 9140 | 213026_at | -0.1172 | 0.5207 | |
| GSE29001 | ATG12 | 9140 | 213026_at | 0.1697 | 0.6537 | |
| GSE38129 | ATG12 | 9140 | 213026_at | -0.1784 | 0.1329 | |
| GSE45670 | ATG12 | 9140 | 213026_at | 0.0037 | 0.9856 | |
| GSE53622 | ATG12 | 9140 | 22549 | 0.1030 | 0.0730 | |
| GSE53624 | ATG12 | 9140 | 22549 | 0.2634 | 0.0057 | |
| GSE63941 | ATG12 | 9140 | 213026_at | -1.1298 | 0.0254 | |
| GSE77861 | ATG12 | 9140 | 213026_at | 0.1948 | 0.5474 | |
| GSE97050 | ATG12 | 9140 | A_23_P20970 | -0.0477 | 0.8298 | |
| SRP007169 | ATG12 | 9140 | RNAseq | -0.5596 | 0.1094 | |
| SRP008496 | ATG12 | 9140 | RNAseq | -0.2677 | 0.3128 | |
| SRP064894 | ATG12 | 9140 | RNAseq | 0.0228 | 0.7992 | |
| SRP133303 | ATG12 | 9140 | RNAseq | 0.3823 | 0.0216 | |
| SRP159526 | ATG12 | 9140 | RNAseq | -0.4177 | 0.0847 | |
| SRP193095 | ATG12 | 9140 | RNAseq | -0.3943 | 0.0000 | |
| SRP219564 | ATG12 | 9140 | RNAseq | -0.2969 | 0.2264 | |
| TCGA | ATG12 | 9140 | RNAseq | -0.0336 | 0.5407 |
Upregulated datasets: 0; Downregulated datasets: 1.
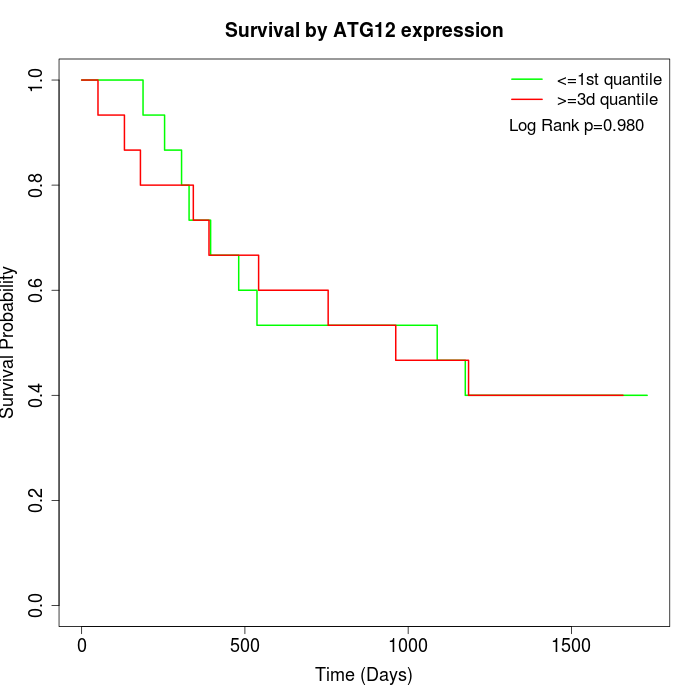
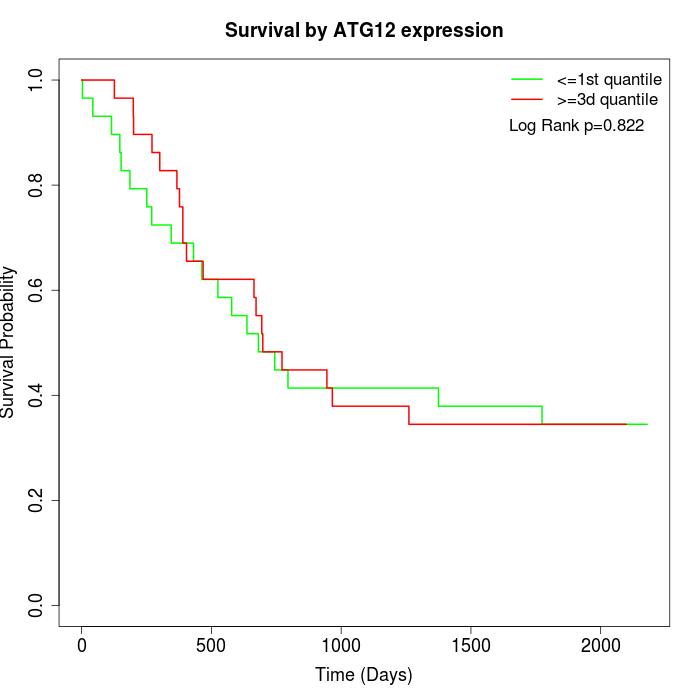
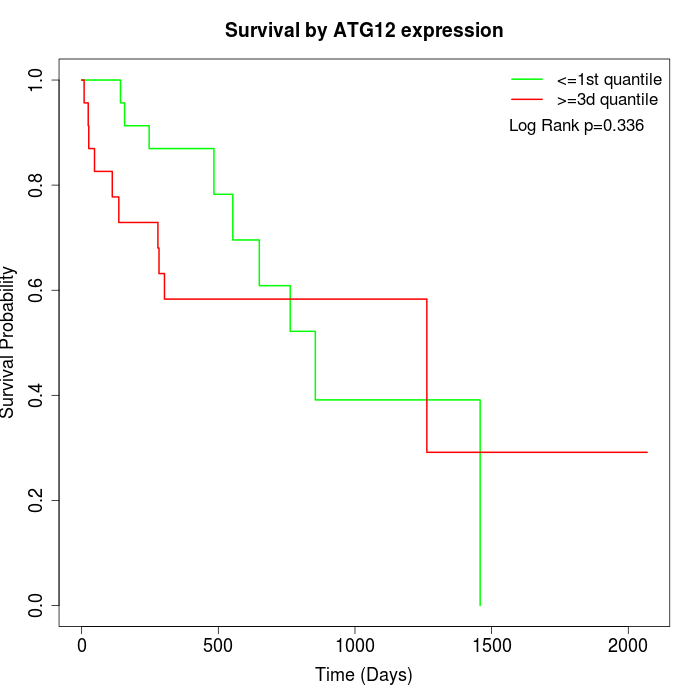
Survival by ATG12 expression:
Note: Click image to view full size file.
Copy number change of ATG12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATG12 | 9140 | 1 | 13 | 16 | |
| GSE20123 | ATG12 | 9140 | 1 | 13 | 16 | |
| GSE43470 | ATG12 | 9140 | 3 | 8 | 32 | |
| GSE46452 | ATG12 | 9140 | 0 | 28 | 31 | |
| GSE47630 | ATG12 | 9140 | 0 | 21 | 19 | |
| GSE54993 | ATG12 | 9140 | 9 | 1 | 60 | |
| GSE54994 | ATG12 | 9140 | 1 | 15 | 37 | |
| GSE60625 | ATG12 | 9140 | 0 | 0 | 11 | |
| GSE74703 | ATG12 | 9140 | 2 | 6 | 28 | |
| GSE74704 | ATG12 | 9140 | 1 | 7 | 12 | |
| TCGA | ATG12 | 9140 | 2 | 41 | 53 |
Total number of gains: 20; Total number of losses: 153; Total Number of normals: 315.
Somatic mutations of ATG12:
Generating mutation plots.
Highly correlated genes for ATG12:
Showing top 20/292 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATG12 | SEC22A | 0.789042 | 3 | 0 | 3 |
| ATG12 | CETN3 | 0.785688 | 3 | 0 | 3 |
| ATG12 | GPCPD1 | 0.75929 | 3 | 0 | 3 |
| ATG12 | HSPA9 | 0.754271 | 3 | 0 | 3 |
| ATG12 | LYSMD3 | 0.748795 | 3 | 0 | 3 |
| ATG12 | CXCL5 | 0.735003 | 3 | 0 | 3 |
| ATG12 | ANKHD1 | 0.732422 | 4 | 0 | 4 |
| ATG12 | EIF4G2 | 0.727825 | 3 | 0 | 3 |
| ATG12 | CCNC | 0.725132 | 4 | 0 | 3 |
| ATG12 | AFF4 | 0.719751 | 4 | 0 | 4 |
| ATG12 | THUMPD3 | 0.719192 | 3 | 0 | 3 |
| ATG12 | IGDCC4 | 0.715003 | 3 | 0 | 3 |
| ATG12 | AIMP1 | 0.714447 | 4 | 0 | 3 |
| ATG12 | DPY19L3 | 0.713978 | 3 | 0 | 3 |
| ATG12 | RNF145 | 0.712074 | 4 | 0 | 3 |
| ATG12 | AP3S1 | 0.711039 | 7 | 0 | 6 |
| ATG12 | ZNF441 | 0.70893 | 3 | 0 | 3 |
| ATG12 | PGK1 | 0.708433 | 3 | 0 | 3 |
| ATG12 | MANEA | 0.702198 | 3 | 0 | 3 |
| ATG12 | MTIF2 | 0.697458 | 3 | 0 | 3 |
For details and further investigation, click here