| Full name: inositol polyphosphate phosphatase like 1 | Alias Symbol: SHIP2 | ||
| Type: protein-coding gene | Cytoband: 11q13.4 | ||
| Entrez ID: 3636 | HGNC ID: HGNC:6080 | Ensembl Gene: ENSG00000165458 | OMIM ID: 600829 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
INPPL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04910 | Insulin signaling pathway |
Expression of INPPL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INPPL1 | 3636 | 201598_s_at | 0.8466 | 0.0939 | |
| GSE20347 | INPPL1 | 3636 | 201598_s_at | 0.4501 | 0.0014 | |
| GSE23400 | INPPL1 | 3636 | 201598_s_at | 0.2744 | 0.0000 | |
| GSE26886 | INPPL1 | 3636 | 201598_s_at | 0.3770 | 0.0684 | |
| GSE29001 | INPPL1 | 3636 | 201598_s_at | 0.2674 | 0.3474 | |
| GSE38129 | INPPL1 | 3636 | 201598_s_at | 0.3102 | 0.0856 | |
| GSE45670 | INPPL1 | 3636 | 201598_s_at | 0.2224 | 0.2360 | |
| GSE53622 | INPPL1 | 3636 | 35228 | 0.2700 | 0.0005 | |
| GSE53624 | INPPL1 | 3636 | 35228 | 0.2733 | 0.0004 | |
| GSE63941 | INPPL1 | 3636 | 201598_s_at | -0.6960 | 0.2283 | |
| GSE77861 | INPPL1 | 3636 | 201598_s_at | 0.2827 | 0.1522 | |
| GSE97050 | INPPL1 | 3636 | A_33_P3358253 | 0.0809 | 0.7841 | |
| SRP007169 | INPPL1 | 3636 | RNAseq | 0.8547 | 0.0535 | |
| SRP008496 | INPPL1 | 3636 | RNAseq | 0.7754 | 0.0037 | |
| SRP064894 | INPPL1 | 3636 | RNAseq | 0.4748 | 0.0116 | |
| SRP133303 | INPPL1 | 3636 | RNAseq | 0.7583 | 0.0034 | |
| SRP159526 | INPPL1 | 3636 | RNAseq | -0.0160 | 0.9562 | |
| SRP193095 | INPPL1 | 3636 | RNAseq | 0.6084 | 0.0014 | |
| SRP219564 | INPPL1 | 3636 | RNAseq | 0.0901 | 0.7803 | |
| TCGA | INPPL1 | 3636 | RNAseq | -0.1344 | 0.0466 |
Upregulated datasets: 0; Downregulated datasets: 0.
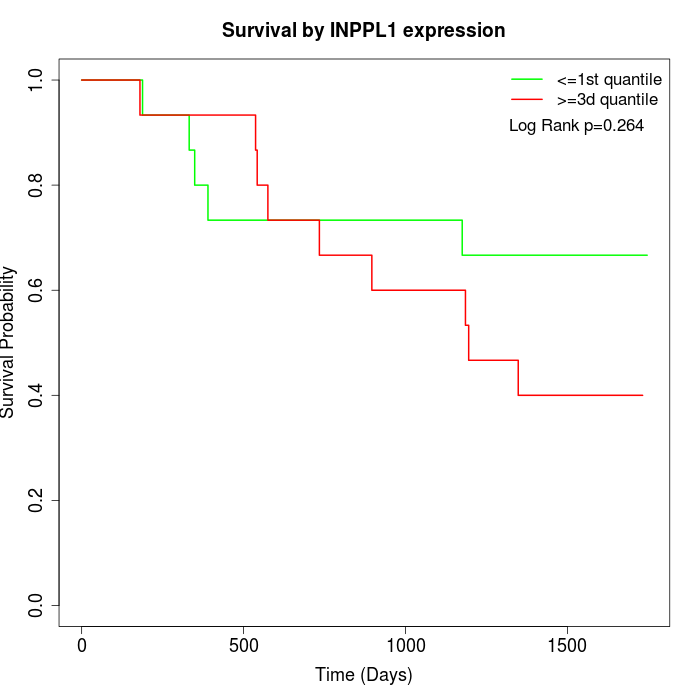
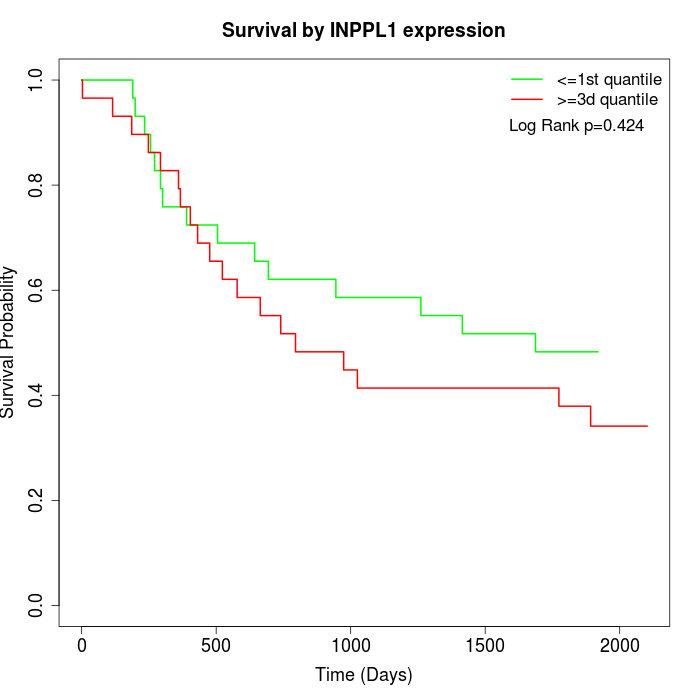
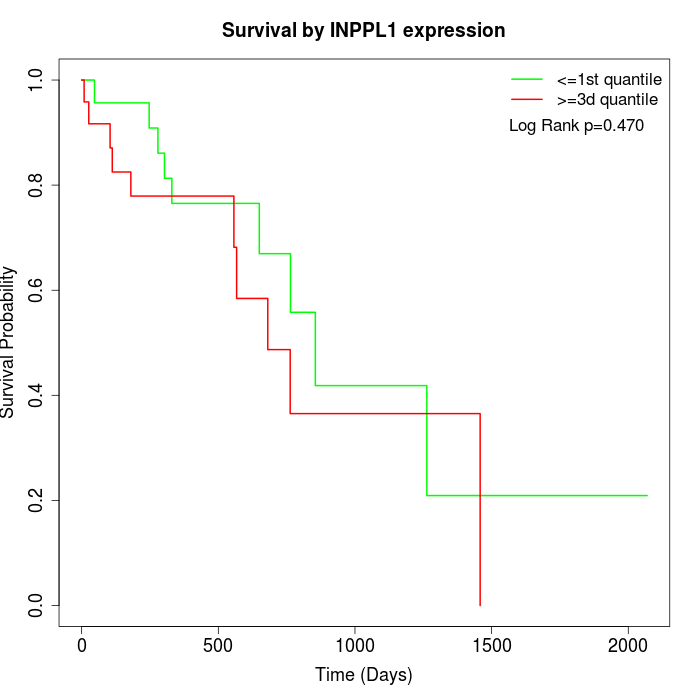
Survival by INPPL1 expression:
Note: Click image to view full size file.
Copy number change of INPPL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INPPL1 | 3636 | 10 | 7 | 13 | |
| GSE20123 | INPPL1 | 3636 | 10 | 7 | 13 | |
| GSE43470 | INPPL1 | 3636 | 9 | 4 | 30 | |
| GSE46452 | INPPL1 | 3636 | 12 | 17 | 30 | |
| GSE47630 | INPPL1 | 3636 | 10 | 10 | 20 | |
| GSE54993 | INPPL1 | 3636 | 7 | 3 | 60 | |
| GSE54994 | INPPL1 | 3636 | 11 | 13 | 29 | |
| GSE60625 | INPPL1 | 3636 | 0 | 7 | 4 | |
| GSE74703 | INPPL1 | 3636 | 7 | 3 | 26 | |
| GSE74704 | INPPL1 | 3636 | 7 | 4 | 9 | |
| TCGA | INPPL1 | 3636 | 35 | 20 | 41 |
Total number of gains: 118; Total number of losses: 95; Total Number of normals: 275.
Somatic mutations of INPPL1:
Generating mutation plots.
Highly correlated genes for INPPL1:
Showing top 20/197 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INPPL1 | TAS2R31 | 0.696789 | 3 | 0 | 3 |
| INPPL1 | F2RL3 | 0.680809 | 3 | 0 | 3 |
| INPPL1 | TMEM235 | 0.644269 | 3 | 0 | 3 |
| INPPL1 | EBF4 | 0.64225 | 3 | 0 | 3 |
| INPPL1 | HIF1AN | 0.641065 | 3 | 0 | 3 |
| INPPL1 | KRTAP10-12 | 0.636239 | 3 | 0 | 3 |
| INPPL1 | SUPV3L1 | 0.635417 | 3 | 0 | 3 |
| INPPL1 | CDH24 | 0.635199 | 3 | 0 | 3 |
| INPPL1 | CASC15 | 0.632483 | 3 | 0 | 3 |
| INPPL1 | C9orf153 | 0.632329 | 3 | 0 | 3 |
| INPPL1 | HEATR6 | 0.631009 | 4 | 0 | 4 |
| INPPL1 | C2CD4D | 0.626316 | 3 | 0 | 3 |
| INPPL1 | RHBDD3 | 0.618235 | 6 | 0 | 5 |
| INPPL1 | C19orf71 | 0.613612 | 3 | 0 | 3 |
| INPPL1 | FOXE3 | 0.612618 | 3 | 0 | 3 |
| INPPL1 | TMEM179 | 0.608607 | 3 | 0 | 3 |
| INPPL1 | RPRML | 0.607532 | 3 | 0 | 3 |
| INPPL1 | FBXO44 | 0.605456 | 6 | 0 | 4 |
| INPPL1 | SIPA1L3 | 0.604554 | 4 | 0 | 4 |
| INPPL1 | PAX6 | 0.603573 | 4 | 0 | 3 |
For details and further investigation, click here