| Full name: inversin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q31.1 | ||
| Entrez ID: 27130 | HGNC ID: HGNC:17870 | Ensembl Gene: ENSG00000119509 | OMIM ID: 243305 |
| Drug and gene relationship at DGIdb | |||
INVS involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04310 | Wnt signaling pathway |
Expression of INVS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INVS | 27130 | 210114_at | 0.0881 | 0.8020 | |
| GSE20347 | INVS | 27130 | 210114_at | 0.1529 | 0.4321 | |
| GSE23400 | INVS | 27130 | 210114_at | 0.2174 | 0.0004 | |
| GSE26886 | INVS | 27130 | 210114_at | 1.3150 | 0.0000 | |
| GSE29001 | INVS | 27130 | 210114_at | 0.3223 | 0.3862 | |
| GSE38129 | INVS | 27130 | 210114_at | 0.2018 | 0.1797 | |
| GSE45670 | INVS | 27130 | 210114_at | 0.6488 | 0.0001 | |
| GSE53622 | INVS | 27130 | 97086 | 0.2187 | 0.0015 | |
| GSE53624 | INVS | 27130 | 97086 | 0.4409 | 0.0000 | |
| GSE63941 | INVS | 27130 | 210114_at | 0.1022 | 0.8342 | |
| GSE77861 | INVS | 27130 | 210114_at | 0.2160 | 0.3033 | |
| GSE97050 | INVS | 27130 | A_23_P157970 | -0.4950 | 0.1658 | |
| SRP007169 | INVS | 27130 | RNAseq | 0.7899 | 0.0529 | |
| SRP008496 | INVS | 27130 | RNAseq | 0.7672 | 0.0051 | |
| SRP064894 | INVS | 27130 | RNAseq | 0.4623 | 0.0089 | |
| SRP133303 | INVS | 27130 | RNAseq | 0.0974 | 0.5563 | |
| SRP159526 | INVS | 27130 | RNAseq | 0.3689 | 0.1995 | |
| SRP193095 | INVS | 27130 | RNAseq | 0.3545 | 0.0006 | |
| SRP219564 | INVS | 27130 | RNAseq | 0.0153 | 0.9528 | |
| TCGA | INVS | 27130 | RNAseq | 0.0374 | 0.5968 |
Upregulated datasets: 1; Downregulated datasets: 0.
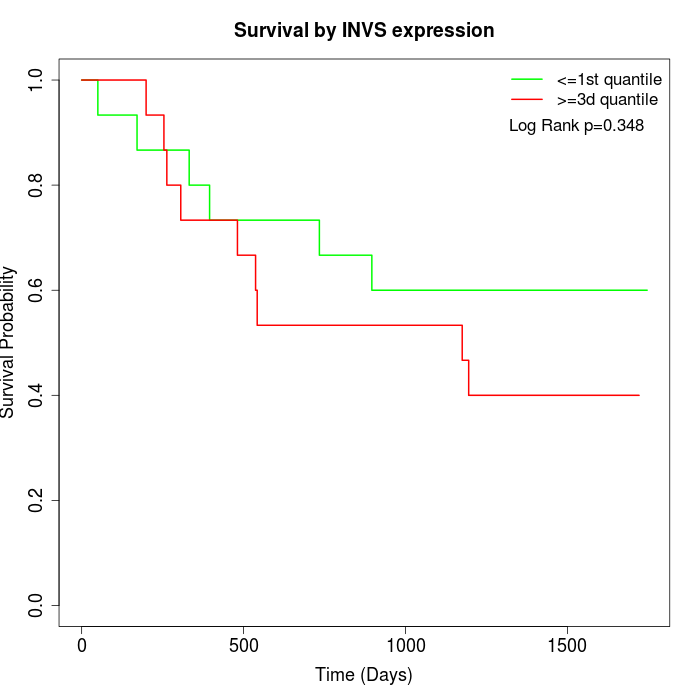
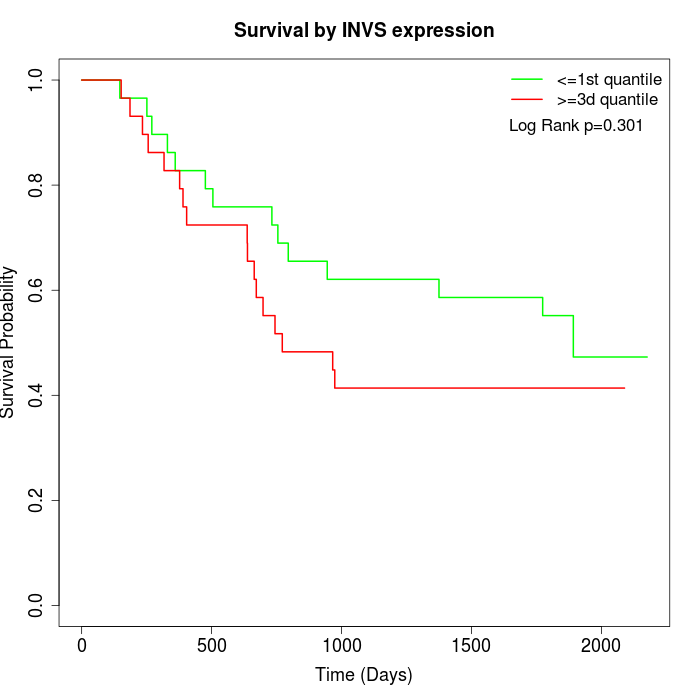
Survival by INVS expression:
Note: Click image to view full size file.
Copy number change of INVS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INVS | 27130 | 4 | 9 | 17 | |
| GSE20123 | INVS | 27130 | 5 | 9 | 16 | |
| GSE43470 | INVS | 27130 | 6 | 3 | 34 | |
| GSE46452 | INVS | 27130 | 6 | 13 | 40 | |
| GSE47630 | INVS | 27130 | 1 | 17 | 22 | |
| GSE54993 | INVS | 27130 | 3 | 3 | 64 | |
| GSE54994 | INVS | 27130 | 9 | 11 | 33 | |
| GSE60625 | INVS | 27130 | 0 | 0 | 11 | |
| GSE74703 | INVS | 27130 | 5 | 3 | 28 | |
| GSE74704 | INVS | 27130 | 2 | 6 | 12 | |
| TCGA | INVS | 27130 | 27 | 22 | 47 |
Total number of gains: 68; Total number of losses: 96; Total Number of normals: 324.
Somatic mutations of INVS:
Generating mutation plots.
Highly correlated genes for INVS:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INVS | BIRC6 | 0.801665 | 3 | 0 | 3 |
| INVS | TWIST2 | 0.785745 | 3 | 0 | 3 |
| INVS | USP40 | 0.770759 | 3 | 0 | 3 |
| INVS | SDR39U1 | 0.769701 | 3 | 0 | 3 |
| INVS | FAM168A | 0.766897 | 3 | 0 | 3 |
| INVS | FAF2 | 0.732913 | 5 | 0 | 5 |
| INVS | HERPUD2 | 0.724816 | 3 | 0 | 3 |
| INVS | SERPINA3 | 0.723529 | 3 | 0 | 3 |
| INVS | PPP6R3 | 0.72132 | 3 | 0 | 3 |
| INVS | IDS | 0.71901 | 3 | 0 | 3 |
| INVS | AMDHD1 | 0.716026 | 3 | 0 | 3 |
| INVS | MTX3 | 0.707706 | 3 | 0 | 3 |
| INVS | MB21D2 | 0.691699 | 4 | 0 | 3 |
| INVS | CERCAM | 0.691113 | 4 | 0 | 4 |
| INVS | TLK1 | 0.684199 | 3 | 0 | 3 |
| INVS | SEMA3C | 0.68313 | 3 | 0 | 3 |
| INVS | ANKRD9 | 0.675839 | 4 | 0 | 3 |
| INVS | NGF | 0.67118 | 3 | 0 | 3 |
| INVS | SPIRE1 | 0.670453 | 5 | 0 | 3 |
| INVS | PTGS2 | 0.667963 | 3 | 0 | 3 |
For details and further investigation, click here