| Full name: JunD proto-oncogene, AP-1 transcription factor subunit | Alias Symbol: AP-1 | ||
| Type: protein-coding gene | Cytoband: 19p13.11 | ||
| Entrez ID: 3727 | HGNC ID: HGNC:6206 | Ensembl Gene: ENSG00000130522 | OMIM ID: 165162 |
| Drug and gene relationship at DGIdb | |||
JUND involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04380 | Osteoclast differentiation |
Expression of JUND:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | JUND | 3727 | 203752_s_at | -0.2619 | 0.6697 | |
| GSE20347 | JUND | 3727 | 203752_s_at | -0.3315 | 0.0403 | |
| GSE23400 | JUND | 3727 | 203752_s_at | -0.3844 | 0.0000 | |
| GSE26886 | JUND | 3727 | 203752_s_at | 0.1016 | 0.6988 | |
| GSE29001 | JUND | 3727 | 203752_s_at | -0.4477 | 0.2653 | |
| GSE38129 | JUND | 3727 | 203752_s_at | -0.5890 | 0.0004 | |
| GSE45670 | JUND | 3727 | 203752_s_at | -0.7414 | 0.0003 | |
| GSE53622 | JUND | 3727 | 45627 | -0.8556 | 0.0000 | |
| GSE53624 | JUND | 3727 | 45627 | -0.5904 | 0.0000 | |
| GSE63941 | JUND | 3727 | 203752_s_at | 0.1115 | 0.8805 | |
| GSE77861 | JUND | 3727 | 203752_s_at | 0.4146 | 0.1869 | |
| GSE97050 | JUND | 3727 | A_33_P3324909 | -0.3683 | 0.5151 | |
| SRP007169 | JUND | 3727 | RNAseq | -1.3781 | 0.0035 | |
| SRP008496 | JUND | 3727 | RNAseq | -1.2703 | 0.0000 | |
| SRP064894 | JUND | 3727 | RNAseq | -0.1835 | 0.5837 | |
| SRP133303 | JUND | 3727 | RNAseq | -0.8035 | 0.0004 | |
| SRP159526 | JUND | 3727 | RNAseq | -0.1666 | 0.4987 | |
| SRP193095 | JUND | 3727 | RNAseq | -0.0732 | 0.7254 | |
| SRP219564 | JUND | 3727 | RNAseq | -0.1043 | 0.8572 | |
| TCGA | JUND | 3727 | RNAseq | -0.2178 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 2.
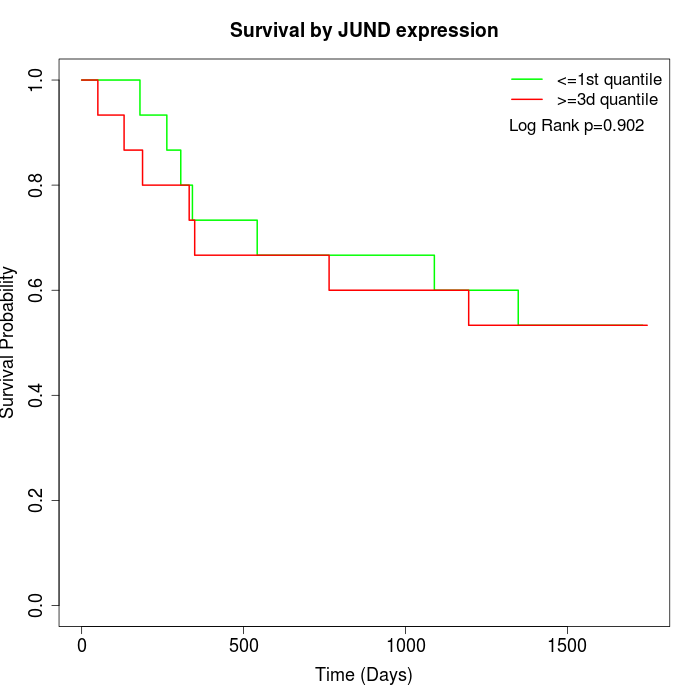
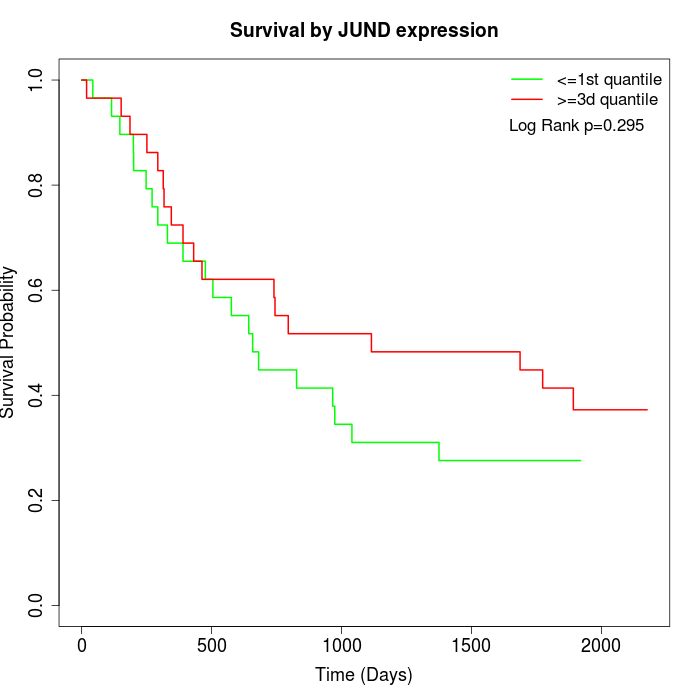
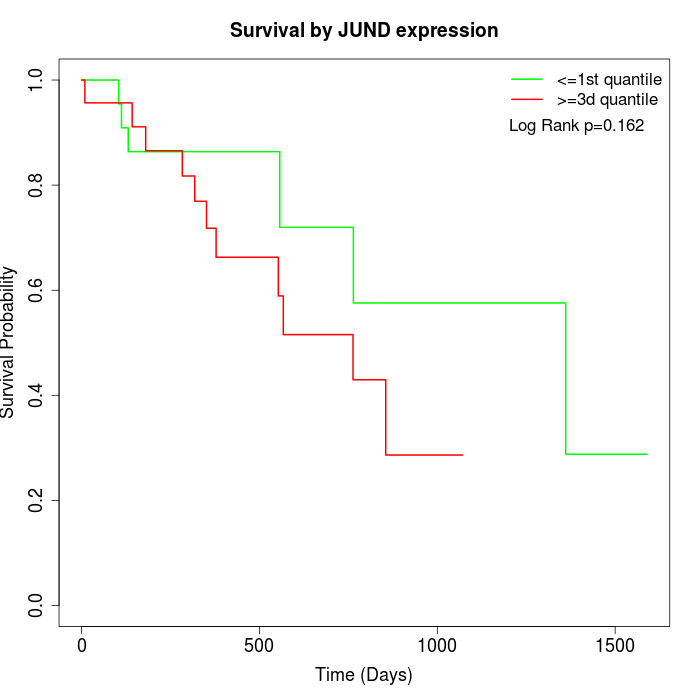
Survival by JUND expression:
Note: Click image to view full size file.
Copy number change of JUND:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | JUND | 3727 | 4 | 4 | 22 | |
| GSE20123 | JUND | 3727 | 3 | 2 | 25 | |
| GSE43470 | JUND | 3727 | 2 | 6 | 35 | |
| GSE46452 | JUND | 3727 | 47 | 1 | 11 | |
| GSE47630 | JUND | 3727 | 4 | 8 | 28 | |
| GSE54993 | JUND | 3727 | 15 | 4 | 51 | |
| GSE54994 | JUND | 3727 | 6 | 13 | 34 | |
| GSE60625 | JUND | 3727 | 9 | 0 | 2 | |
| GSE74703 | JUND | 3727 | 2 | 4 | 30 | |
| GSE74704 | JUND | 3727 | 0 | 2 | 18 | |
| TCGA | JUND | 3727 | 16 | 10 | 70 |
Total number of gains: 108; Total number of losses: 54; Total Number of normals: 326.
Somatic mutations of JUND:
Generating mutation plots.
Highly correlated genes for JUND:
Showing top 20/328 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| JUND | CCNL1 | 0.76292 | 3 | 0 | 3 |
| JUND | ZNF609 | 0.752174 | 3 | 0 | 3 |
| JUND | SERTAD1 | 0.745351 | 4 | 0 | 4 |
| JUND | NRL | 0.726899 | 3 | 0 | 3 |
| JUND | IRF2BP2 | 0.717576 | 3 | 0 | 3 |
| JUND | CSRNP1 | 0.705677 | 6 | 0 | 6 |
| JUND | GPS2 | 0.70479 | 3 | 0 | 3 |
| JUND | SLC25A25 | 0.698026 | 5 | 0 | 5 |
| JUND | EGFL7 | 0.687595 | 3 | 0 | 3 |
| JUND | ISM2 | 0.681055 | 3 | 0 | 3 |
| JUND | DUSP2 | 0.679505 | 4 | 0 | 3 |
| JUND | CBX4 | 0.663093 | 3 | 0 | 3 |
| JUND | FOSB | 0.659481 | 12 | 0 | 11 |
| JUND | TMEM150A | 0.6591 | 3 | 0 | 3 |
| JUND | RILPL1 | 0.658902 | 4 | 0 | 3 |
| JUND | DDX42 | 0.65823 | 3 | 0 | 3 |
| JUND | EMD | 0.655668 | 5 | 0 | 4 |
| JUND | INO80E | 0.654119 | 3 | 0 | 3 |
| JUND | MAT2A | 0.645156 | 4 | 0 | 3 |
| JUND | GH2 | 0.644671 | 3 | 0 | 3 |
For details and further investigation, click here