| Full name: growth hormone 2 | Alias Symbol: GH-V|GHV|GHL|hGH-V | ||
| Type: protein-coding gene | Cytoband: 17q23.3 | ||
| Entrez ID: 2689 | HGNC ID: HGNC:4262 | Ensembl Gene: ENSG00000136487 | OMIM ID: 139240 |
| Drug and gene relationship at DGIdb | |||
GH2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04630 | Jak-STAT signaling pathway |
Expression of GH2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GH2 | 2689 | 206195_x_at | -0.1572 | 0.4004 | |
| GSE20347 | GH2 | 2689 | 206195_x_at | -0.0722 | 0.3440 | |
| GSE23400 | GH2 | 2689 | 206195_x_at | -0.0676 | 0.0384 | |
| GSE26886 | GH2 | 2689 | 206195_x_at | -0.1124 | 0.3501 | |
| GSE29001 | GH2 | 2689 | 206195_x_at | -0.2626 | 0.0725 | |
| GSE38129 | GH2 | 2689 | 206195_x_at | -0.1234 | 0.1470 | |
| GSE45670 | GH2 | 2689 | 206195_x_at | -0.0411 | 0.6124 | |
| GSE53622 | GH2 | 2689 | 114898 | 0.0962 | 0.5539 | |
| GSE53624 | GH2 | 2689 | 114898 | -0.1717 | 0.1282 | |
| GSE63941 | GH2 | 2689 | 206195_x_at | -0.0514 | 0.7100 | |
| GSE77861 | GH2 | 2689 | 206195_x_at | -0.1416 | 0.2865 | |
| GSE97050 | GH2 | 2689 | A_23_P207174 | -0.2439 | 0.2371 |
Upregulated datasets: 0; Downregulated datasets: 0.
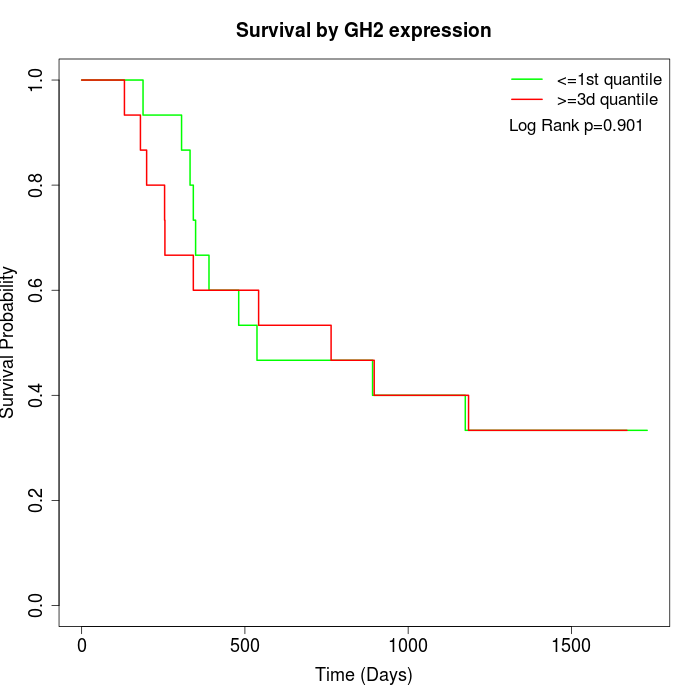
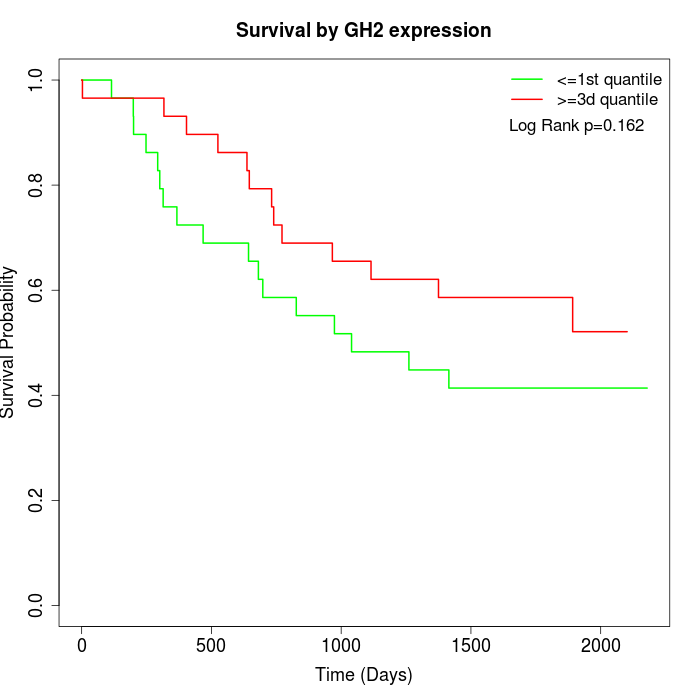
Survival by GH2 expression:
Note: Click image to view full size file.
Copy number change of GH2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GH2 | 2689 | 3 | 1 | 26 | |
| GSE20123 | GH2 | 2689 | 3 | 1 | 26 | |
| GSE43470 | GH2 | 2689 | 4 | 0 | 39 | |
| GSE46452 | GH2 | 2689 | 31 | 0 | 28 | |
| GSE47630 | GH2 | 2689 | 7 | 1 | 32 | |
| GSE54993 | GH2 | 2689 | 2 | 5 | 63 | |
| GSE54994 | GH2 | 2689 | 9 | 4 | 40 | |
| GSE60625 | GH2 | 2689 | 4 | 0 | 7 | |
| GSE74703 | GH2 | 2689 | 4 | 0 | 32 | |
| GSE74704 | GH2 | 2689 | 3 | 1 | 16 | |
| TCGA | GH2 | 2689 | 30 | 8 | 58 |
Total number of gains: 100; Total number of losses: 21; Total Number of normals: 367.
Somatic mutations of GH2:
Generating mutation plots.
Highly correlated genes for GH2:
Showing top 20/942 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GH2 | NDUFAF3 | 0.800152 | 3 | 0 | 3 |
| GH2 | PLEKHA5 | 0.778077 | 3 | 0 | 3 |
| GH2 | CGA | 0.770189 | 4 | 0 | 4 |
| GH2 | CLCC1 | 0.764602 | 3 | 0 | 3 |
| GH2 | HSPB6 | 0.745615 | 4 | 0 | 4 |
| GH2 | ADD1 | 0.744312 | 3 | 0 | 3 |
| GH2 | KRTAP5-8 | 0.738217 | 7 | 0 | 7 |
| GH2 | NHSL2 | 0.736339 | 3 | 0 | 3 |
| GH2 | PIGP | 0.733342 | 3 | 0 | 3 |
| GH2 | STOML1 | 0.730219 | 3 | 0 | 3 |
| GH2 | CRHR2 | 0.729025 | 4 | 0 | 3 |
| GH2 | COMMD1 | 0.727356 | 3 | 0 | 3 |
| GH2 | C9orf16 | 0.718152 | 3 | 0 | 3 |
| GH2 | ADH1A | 0.715612 | 4 | 0 | 3 |
| GH2 | TMEM190 | 0.714389 | 3 | 0 | 3 |
| GH2 | ZNF614 | 0.71294 | 3 | 0 | 3 |
| GH2 | FLII | 0.71102 | 3 | 0 | 3 |
| GH2 | RHAG | 0.709722 | 4 | 0 | 4 |
| GH2 | ATN1 | 0.709195 | 4 | 0 | 3 |
| GH2 | LENEP | 0.708246 | 3 | 0 | 3 |
For details and further investigation, click here